3ZOE
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, P-HYDROXYBENZALDEHYDE | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOD
 
 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound benzene-1,4-diol | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, benzene-1,4-diol | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOC
 
 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, P-HYDROXYBENZALDEHYDE | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
8YHR
 
 | | DHODH in complex with furocoumavirin | | Descriptor: | 4-methyl-8-[(S)-oxidanyl(phenyl)methyl]-9-phenyl-furo[2,3-h]chromen-2-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Hara, K, Okumura, H, Nakahara, M, Sato, M, Hashimoto, H, Osada, H, Watanabe, K. | | Deposit date: | 2024-02-28 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analyses of Inhibition of Human Dihydroorotate Dehydrogenase by Antiviral Furocoumavirin.
Biochemistry, 63, 2024
|
|
8Y2S
 
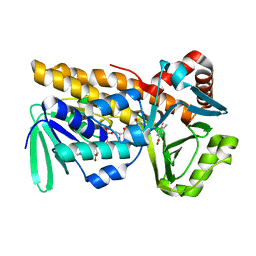 | | P-hydroxybenzoate hydroxylase complexed with 4-hydroxy-3-methylbenzoic acid | | Descriptor: | 3-methyl-4-oxidanyl-benzoic acid, 4-hydroxybenzoate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2024-01-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Enhancement of Flavin-Containing Monooxygenase through Machine Learning Methodology
Acs Catalysis, 14, 2024
|
|
1GS9
 
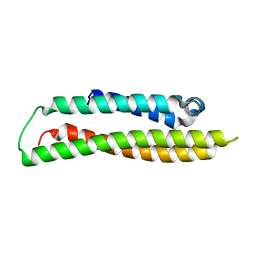 | | Apolipoprotein E4, 22k domain | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Verderame, J.R, Kantardjieff, K, Segelke, B, Weisgraber, K, Rupp, B. | | Deposit date: | 2002-01-02 | | Release date: | 2003-06-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the 22K Domain of Human Apolipoprotein E4
To be Published
|
|
6LL8
 
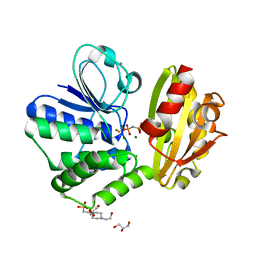 | | Type II inorganic pyrophosphatase (PPase) from the psychrophilic bacterium Shewanella sp. AS-11, Mg-PNP form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, FLUORIDE ION, ... | | Authors: | Horitani, M, Kusubayashi, K, Oshima, K, Yato, A, Sugimoto, H, Watanabe, K. | | Deposit date: | 2019-12-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystallography and Electron Paramagnetic Resonance Spectroscopy Reveal Active Site Rearrangement of Cold-Adapted Inorganic Pyrophosphatase.
Sci Rep, 10, 2020
|
|
6LL7
 
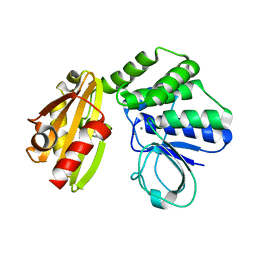 | | Type II inorganic pyrophosphatase (PPase) from the psychrophilic bacterium Shewanella sp. AS-11, Mn-activated form | | Descriptor: | CALCIUM ION, Inorganic pyrophosphatase, MANGANESE (II) ION | | Authors: | Horitani, M, Kusubayashi, K, Oshima, K, Yato, A, Sugimoto, H, Watanabe, K. | | Deposit date: | 2019-12-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Crystallography and Electron Paramagnetic Resonance Spectroscopy Reveal Active Site Rearrangement of Cold-Adapted Inorganic Pyrophosphatase.
Sci Rep, 10, 2020
|
|
2PI3
 
 | |
6LA0
 
 | | Crystal structure of AoRut | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoside hydrolase family 5 | | Authors: | Koseki, T, Makabe, K. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aspergillus oryzae Rutinosidase: Biochemical and Structural Investigation.
Appl.Environ.Microbiol., 87, 2021
|
|
4ETK
 
 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ESS
 
 | | Crystal Structure of E6D/L155R variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR187 | | Descriptor: | OR187 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9971 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETJ
 
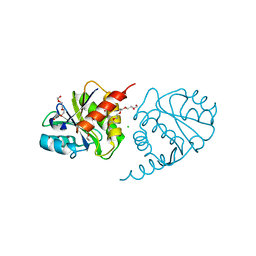 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
7F4B
 
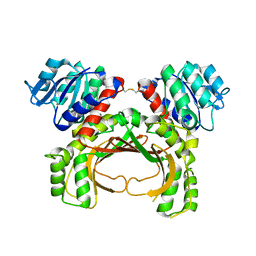 | | The crystal structure of the immature apo-enzyme of homoserine dehydrogenase from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | MAGNESIUM ION, homoserine dehydrogenase | | Authors: | Kurihara, E, Kubota, T, Watanabe, K, Ogata, K, Kaneko, R, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
7F4C
 
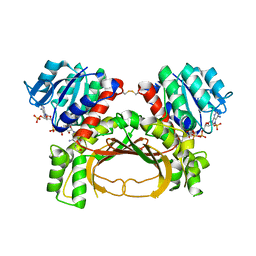 | | The crystal structure of the immature holo-enzyme of homoserine dehydrogenase complexed with NADP and 1,4-butandiol from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | 1,4-BUTANEDIOL, Homoserine dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ogata, K, Kaneko, R, Kubota, T, Watanabe, K, Kurihara, E, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
5YY2
 
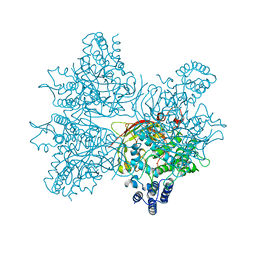 | | Crystal structure of AsqI with Zn | | Descriptor: | Uncharacterized protein AsqI, ZINC ION | | Authors: | Hara, K, Hashimoto, H, Kishimoto, S, Watanabe, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Enzymatic one-step ring contraction for quinolone biosynthesis.
Nat Commun, 9, 2018
|
|
7CP6
 
 | | Crystal structure of FqzB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase, ... | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
5YY3
 
 | |
7CP7
 
 | | Crystal structure of FqzB, native proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
1NGU
 
 | | NMR Structure of Putative 3'Terminator for B. Anthracis pagA Gene Noncoding Strand | | Descriptor: | 5'-D(*CP*TP*CP*TP*CP*CP*TP*TP*GP*TP*AP*TP*TP*TP*CP*TP*TP*AP*CP*AP*AP*AP*AP*AP*GP*AP*G)-3' | | Authors: | Shiflett, P.R, Taylor-McCabe, K.J, Michalczyk, R, Silks, L.A, Gupta, G. | | Deposit date: | 2002-12-17 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on the Hairpins at the 3' Untranslated Region of an Anthrax Toxin Gene
Biochemistry, 42, 2003
|
|
6KAW
 
 | | Crystal structure of CghA | | Descriptor: | CghA | | Authors: | Hara, K, Hashimoto, H, Yokoyama, M, Sato, M, Watanabe, K. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels-Alderase
Nat Catal, 2021
|
|
4XK0
 
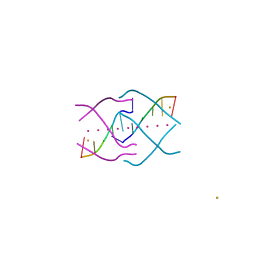 | | Crystal structure of a tetramolecular RNA G-quadruplex in potassium | | Descriptor: | BARIUM ION, POTASSIUM ION, RNA (5'-(*UP*GP*GP*GP*GP*U)-3') | | Authors: | Chen, M.C, Murat, P, Abecassis, K.A, Ferre-D'Amare, A.R, Balasubramanian, S. | | Deposit date: | 2015-01-09 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Insights into the mechanism of a G-quadruplex-unwinding DEAH-box helicase.
Nucleic Acids Res., 43, 2015
|
|
6KBC
 
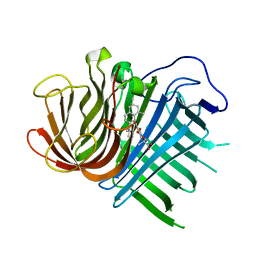 | | Crystal structure of CghA with Sch210972 | | Descriptor: | (2S)-3-[(2S,4E)-4-[[(1R,2S,4aR,6S,8R,8aS)-2-[(E)-but-2-en-2-yl]-6,8-dimethyl-1,2,4a,5,6,7,8,8a-octahydronaphthalen-1-yl]-oxidanyl-methylidene]-3,5-bis(oxidanylidene)pyrrolidin-2-yl]-2-methyl-2-oxidanyl-propanoic acid, CghA | | Authors: | Hara, K, Hashimoto, H, Maeda, N, Sato, M, Watanabe, K. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels-Alderase
Nat Catal, 2021
|
|
6KJI
 
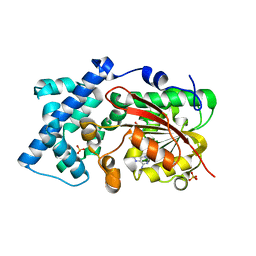 | | Crystal structure of PsoF with SAH | | Descriptor: | Dual-functional monooxygenase/methyltransferase psoF, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analyses oftrans C-Methyltransferase in Fungal Polyketide Biosynthesis.
Biochemistry, 58, 2019
|
|
