4QNN
 
 | |
7TRL
 
 | | Crystal structure of human BIRC2 BIR3 domain in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, Histone H3, ... | | Authors: | Klein, B.J, Tencer, A.H, Kutateladze, T.G. | | Deposit date: | 2022-01-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TRM
 
 | | Crystal structure of human BIRC2 BIR3 domain in complex with inhibitor LCL-161 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, LCL-161, ... | | Authors: | Tencer, A.H, Klein, B.J, Kutateladze, T.G. | | Deposit date: | 2022-01-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7UMJ
 
 | | Crystal structure of recombinant Solieria filiformis lectin (rSfL) | | Descriptor: | Lectin SfL-1, SULFATE ION | | Authors: | Chaves, R.P, Bezerra, E.H.S, da Silva, F.M.S, Carneiro, R.F, Sampaio, A.H, Rocha, B.A.M, Nagano, C.S. | | Deposit date: | 2022-04-07 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural study and antimicrobial and wound healing effects of lectin from Solieria filiformis (Kutzing) P.W.Gabrielson.
Biochimie, 214, 2023
|
|
7CGA
 
 | |
5TW3
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2016-11-11 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural and Preclinical Studies of Computationally Designed Non-Nucleoside Reverse Transcriptase Inhibitors for Treating HIV infection.
Mol. Pharmacol., 91, 2017
|
|
5U38
 
 | | Crystal structure of native lectin from Platypodium elegans seeds (PELa) complexed with Man1-3Man-OMe. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lectin, ... | | Authors: | Silva, I.B, Araripe, D.A, Neco, A.H.B, Pinto-Junior, V.R, Osterne, V.J.S, Santiago, M.Q, Silva-Filho, J.C, Leal, R.B, Rocha, C.R.C, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2016-12-01 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies and nociceptive activity of a native lectin from Platypodium elegans seeds (nPELa).
Int. J. Biol. Macromol., 107, 2018
|
|
5U3E
 
 | | Crystal Structure of Native Lectin from Canavalia bonariensis Seeds (CaBo) complexed with alpha-methyl-D-mannoside | | Descriptor: | CALCIUM ION, Canavalia bonariensis seed lectin, MANGANESE (II) ION, ... | | Authors: | Silva, M.T.L, Osterne, V.J.S, Pinto-Junior, V.R, Santiago, M.Q, Araripe, D.A, Neco, A.H.B, Silva-Filho, J.C, Martins, J.L, Rocha, C.R.C, Leal, R.B, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Canavalia bonariensis lectin: Molecular bases of glycoconjugates interaction and antiglioma potential.
Int. J. Biol. Macromol., 106, 2018
|
|
5TUN
 
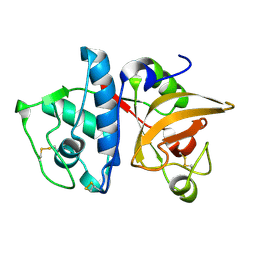 | | Crystal structure of uninhibited human Cathepsin K at 1.62 Angstrom resolution | | Descriptor: | Cathepsin K | | Authors: | Aguda, A.H, Kruglyak, N, Nguyen, N.T, Law, S, Bromme, D, Brayer, G.D. | | Deposit date: | 2016-11-06 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of mouse cathepsin K structural elements that regulate the potency of odanacatib.
Biochem. J., 474, 2017
|
|
4QUN
 
 | |
4QUM
 
 | |
4RH5
 
 | |
4RH9
 
 | |
4RHG
 
 | |
4RI4
 
 | |
4RI5
 
 | |
5TG3
 
 | | Crystal Structure of Dioclea reflexa seed lectin (DrfL) in complex with X-Man | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Dioclea reflexa lectin, ... | | Authors: | Santiago, M.Q, Correia, J.L.A, Pinto-Junior, V.R, Osterne, V.J.S, Pereira, R.I, Silva-Filho, J.C, Lossio, C.F, Rocha, B.A.M, Delatorre, P, Neco, A.H.B, Araripe, D.A, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2016-09-27 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Structural studies of a vasorelaxant lectin from Dioclea reflexa Hook seeds: Crystal structure, molecular docking and dynamics.
Int. J. Biol. Macromol., 98, 2017
|
|
4S0G
 
 | |
5VQW
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acrylamide (JLJ685), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)prop-2-enamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Kudalkar, S.N, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQR
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQZ
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) Variant in Complex with 2-chloro-N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacetamide (JLJ686), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Buckingham, A.B, Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4UFI
 
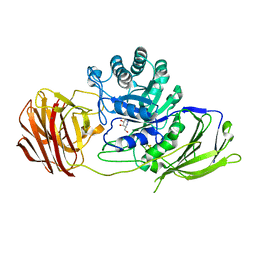 | | Mouse Galactocerebrosidase complexed with aza-galacto-fagomine AGF | | Descriptor: | (3R,4S,5R)-3-(hydroxymethyl)-1,2-diazinane-4,5-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hill, C.H, Viuff, A.H, Spratley, S.J, Salamone, S, Christensen, S.H, Read, R.J, Moriarty, N.W, Jensen, H.H, Deane, J.E. | | Deposit date: | 2015-03-17 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Azasugar Inhibitors as Pharmacological Chaperones for Krabbe Disease.
Chem.Sci., 6, 2015
|
|
5UUY
 
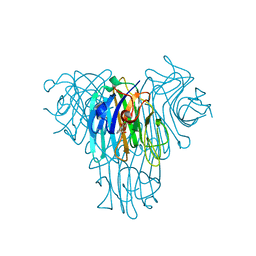 | | Crystal structure of Dioclea lasiocarpa lectin (DLL) complexed with X-MAN | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Dioclea lasiocarpa lectin (DLL), ... | | Authors: | Santiago, M.Q, Pinto-Junior, V.R, Osterne, V.J.S, Rocha, C.R.C, Neco, A.H.B, Fonseca, F.M.P, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2017-02-17 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural analysis of Dioclea lasiocarpa lectin: A C6 cells apoptosis-inducing protein.
Int. J. Biochem. Cell Biol., 92, 2017
|
|
4UFH
 
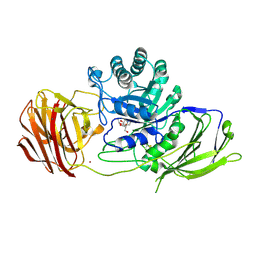 | | Mouse Galactocerebrosidase complexed with iso-galacto-fagomine IGF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hill, C.H, Viuff, A.H, Spratley, S.J, Salamone, S, Christensen, S.H, Read, R.J, Moriarty, N.W, Jensen, H.H, Deane, J.E. | | Deposit date: | 2015-03-17 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Azasugar Inhibitors as Pharmacological Chaperones for Krabbe Disease.
Chem.Sci., 6, 2015
|
|
7D2A
 
 | | CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, AlyQ, CALCIUM ION, ... | | Authors: | Teh, A.H, Sim, P.F. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for binding uronic acids by family 32 carbohydrate-binding modules.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
