6ZBA
 
 | | Crystal structure of PDE4D2 in complex with inhibitor LEO39652 | | Descriptor: | 1,2-ETHANEDIOL, 2-methylpropyl 1-[8-methoxy-5-(1-oxidanylidene-3~{H}-2-benzofuran-5-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]cyclopropane-1-carboxylate, DIMETHYL SULFOXIDE, ... | | Authors: | Akutsu, M, Hakansson, M, Welin, M, Svensson, A, Logan, D.T, Sorensen, M.D. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Early Clinical Development of Isobutyl 1-[8-Methoxy-5-(1-oxo-3 H -isobenzofuran-5-yl)-[1,2,4]triazolo[1,5- a ]pyridin-2-yl]cyclopropanecarboxylate (LEO 39652), a Novel "Dual-Soft" PDE4 Inhibitor for Topical Treatment of Atopic Dermatitis.
J.Med.Chem., 63, 2020
|
|
6R5X
 
 | |
6R64
 
 | | N-terminal domain of modification dependent EcoKMcrA restriction endonuclease (NEco) in complex with C5mCGG target sequence | | Descriptor: | 5-methylcytosine-specific restriction enzyme A, DNA (5'-D(*GP*AP*AP*CP*(5CM)P*GP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*(5CM)P*GP*GP*TP*TP*C)-3') | | Authors: | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
5N0D
 
 | | Crystal structure of human carbonic anhydrase II in complex with (R)-4-(6,7-dihydroxy-1-phenyl-3,4-tetrahydroisoquinoline-1H-2-carbonyl)benzenesulfonamide. | | Descriptor: | (R)-4-(6,7-dihydroxy-1-phenyl-3,4-tetrahydroisoquinoline-1H-2-carbonyl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing Molecular Interactions between Human Carbonic Anhydrases (hCAs) and a Novel Class of Benzenesulfonamides.
J. Med. Chem., 60, 2017
|
|
6XVA
 
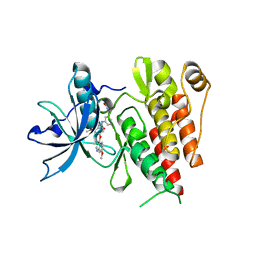 | | Crystal structure of the kinase domain of human c-KIT in complex with a type-II inhibitor bearing an acrylamide | | Descriptor: | Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, ~{N}-[[3-[2-[3-methoxy-5-(7-methoxyquinolin-4-yl)oxy-pyridin-2-yl]ethanoylamino]-5-methyl-phenyl]methyl]propanamide | | Authors: | Schimpl, M, McAuley, K, Hoyt, E.A, Thomas, M, Bodnarchuk, M.S, Lewis, H.J, Barratt, D, Deery, M.J, Ogg, D.J, Bernardes, G.J.L, Ward, R.A, Kettle, J.G, Waring, M.J. | | Deposit date: | 2020-01-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alkynyl Benzoxazines and Dihydroquinazolines as Cysteine Targeting Covalent Warheads and Their Application in Identification of Selective Irreversible Kinase Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
8FU8
 
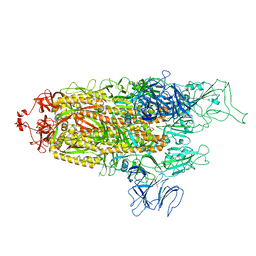 | | Structure of Covid Spike variant deltaN135 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
5N1H
 
 | |
6ZLN
 
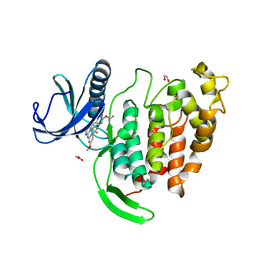 | | CLK1 bound with GW807982X (Cpd 8) | | Descriptor: | 1,2-ETHANEDIOL, 4-(6-ethoxypyrazolo[1,5-b]pyridazin-3-yl)-~{N}-[3-methoxy-5-(trifluoromethyl)phenyl]pyrimidin-2-amine, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
8FU9
 
 | | Structure of Covid Spike variant deltaN25 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
6Z2O
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, O-glycan protease, ZINC ION | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
5JBK
 
 | | Trichoderma harzianum GH1 beta-glucosidase ThBgl1 | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Florindo, R.N, Mutti, H.S, Polikarpov, I, Nascimento, A.S. | | Deposit date: | 2016-04-13 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural insights into beta-glucosidase transglycosylation based on biochemical, structural and computational analysis of two GH1 enzymes from Trichoderma harzianum.
N Biotechnol, 40, 2018
|
|
6Z2V
 
 | | CLK3 A319V mutant bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, ... | | Authors: | Schroeder, M, Chaikuad, A, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
5N6W
 
 | | Retinoschisin R141H Mutant | | Descriptor: | Retinoschisin | | Authors: | Ramsay, E.P, Collins, R.F, Owens, T.W, Siebert, C.A, Jones, R.P.O, Roseman, A, Wang, T, Baldock, C. | | Deposit date: | 2017-02-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of X-linked retinoschisis mutations reveals distinct classes which differentially effect retinoschisin function
Human Molecular Genetics, 25, 2016
|
|
6VIO
 
 | |
4YAV
 
 | | Crystal structure of LigG in complex with B-glutathionyl-acetoveratrone (GS-AV) from Sphingobium sp. strain SYK-6 | | Descriptor: | Glutathione S-transferase, L-gamma-glutamyl-3-{(E)-[2-(3,4-dimethoxyphenyl)-2-oxoethylidene]-lambda~4~-sulfanyl}-L-alanylglycine, SULFATE ION | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
5NGS
 
 | | Crystal structure of human MTH1 in complex with inhibitor 6-[(2-phenylethyl)sulfanyl]-7H-purin-2-amine | | Descriptor: | 6-(2-phenylethylsulfanyl)-7~{H}-purin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
2NWQ
 
 | | Short chain dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | Probable short-chain dehydrogenase, SULFATE ION | | Authors: | McGrath, T.E, Kimber, M.S, Beattie, B, Thambipillai, D, Dharamsi, A, Virag, C, Nethery-Brokx, K, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-11-16 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short chain dehydrogenase from Pseudomonas aeruginosa
To be Published
|
|
8DT9
 
 | | Crystal Structure of SARS CoV-2 Mpro mutant L141R with Pfizer Intravenous Inhibitor PF-00835231 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contributions of Hyperactive Mutations in M pro from SARS-CoV-2 to Drug Resistance.
Acs Infect Dis., 10, 2024
|
|
8E4W
 
 | |
6XPG
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography | | Descriptor: | Lysophospholipase L1 | | Authors: | Kim, Y, Sherrell, D.A, Owen, R, Axford, D, Ebrahim, A, Johnson, J, Welk, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography
To Be Published
|
|
7EV0
 
 | | Crystal structure of pepsin cleaved C-terminal half of lactoferrin at 2.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Singh, J, Ahmad, M.I, Maurya, A, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pepsin cleaved C-terminal half of lactoferrin at 2.7A resolution
To Be Published
|
|
6Z1A
 
 | | Ternary complex of Staphylococcus aureus DNA gyrase with AMK12 and DNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kolaric, A, Germe, T, Hrast, M, Stevenson, C.E.M, Lawson, D.M, Burton, N, Voros, J, Maxwell, A, Minovski, N, Anderluh, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent DNA gyrase inhibitors bind asymmetrically to their target using symmetrical bifurcated halogen bonds.
Nat Commun, 12, 2021
|
|
6OF6
 
 | | Crystal structure of tRNA^ Ala(GGC) bound to cognate 70S A-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Sunita, S, Dunham, C.M. | | Deposit date: | 2019-03-28 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5J10
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | peptide design 2L4HC2_24 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
