8AGC
 
 | | Structure of yeast oligosaccharylransferase complex with lipid-linked oligosaccharide and non-acceptor peptide bound | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramirez, A.S, de Capitani, M, Pesciullesi, G, Kowal, J, Bloch, J.S, Irobalieva, R.N, Aebi, M, Reymond, J.L, Locher, K.P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
4UF0
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with epitherapuetic compound 2-(((2-((2-(dimethylamino)ethyl) (ethyl)amino)-2-oxoethyl)amino)methyl)isonicotinic acid. | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, FE (II) ION, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Tobias, K, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2014-12-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5OKJ
 
 | | Non-conservatively refined structure of Gan1D-WT, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in the C2 spacegroup | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
6MQ3
 
 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Substrate-selective Macrocycle Inhibitor 63 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Insulin-degrading enzyme, {(8R,9S,10S)-9-(2',3'-dimethyl[1,1'-biphenyl]-4-yl)-6-[(1-methyl-1H-imidazol-2-yl)sulfonyl]-1,6-diazabicyclo[6.2.0]decan-10-yl}methanol | | Authors: | Tan, G.A, Seeliger, M.A, Welsh, A.J, Maianti, J.P, Liu, D.R. | | Deposit date: | 2018-10-09 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.569147 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
4UHC
 
 | | Structural studies of a thermophilic esterase from Thermogutta terrifontis (Native) | | Descriptor: | CHLORIDE ION, ESTERASE | | Authors: | Sayer, C, Isupov, M.N, Bonch-Osmolovskaya, E, Littlechild, J.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structural Studies of a Thermophilic Esterase from a New Planctomycetes Species, Thermogutta Terrifontis.
FEBS J., 282, 2015
|
|
7SGD
 
 | | Lassa virus glycoprotein construct(Josiah GPCysR4) recovered from GPC-I53-50 nanoparticle by localized reconstruction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Antanasijevic, A, Brouwer, P.J.M, Ward, A.B. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Lassa virus glycoprotein nanoparticles elicit neutralizing antibody responses and protection.
Cell Host Microbe, 30, 2022
|
|
6FUZ
 
 | | Crystal structure of the TPR domain of KLC1 in complex with the C-terminal peptide of JIP1 | | Descriptor: | GLYCEROL, Kinesin light chain 1,Kinesin light chain 1,C-Jun-amino-terminal kinase-interacting protein 1, nanobody | | Authors: | Pernigo, S, Dodding, M.P, Steiner, R.A. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for isoform-specific kinesin-1 recognition of Y-acidic cargo adaptors.
Elife, 7, 2018
|
|
4UIR
 
 | | Structure of oleate hydratase from Elizabethkingia meningoseptica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HEXAETHYLENE GLYCOL, OLEATE HYDRATASE, ... | | Authors: | Pavkov-Keller, T, Hromic, A, Engleder, M, Emmerstorfer, A, Steinkellner, G, Schrempf, S, Wriessnegger, T, Leitner, E, Strohmeier, G.A, Kaluzna, I, Mink, D, Schuermann, M, Wallner, S, Macheroux, P, Pichler, H, Gruber, K. | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Mechanism of Oleate Hydratase from Elizabethkingia Meningoseptica.
Chembiochem, 16, 2015
|
|
6RV4
 
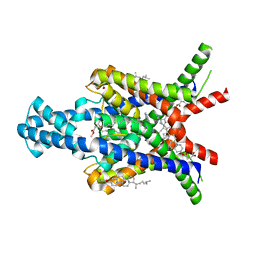 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 2341237 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, POTASSIUM ION, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
5O1G
 
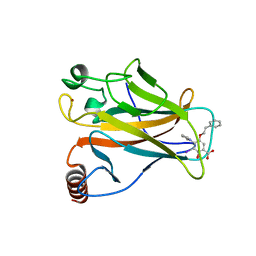 | | p53 cancer mutant Y220C in complex with compound MB487 | | Descriptor: | 3-iodanyl-2-oxidanyl-5-(2-phenylethoxy)-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Baud, M.G.J, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5OG5
 
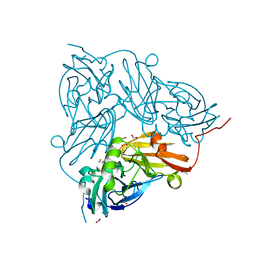 | | Cu nitrite reductase serial data at varying temperatures RT 0.21MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
7OCY
 
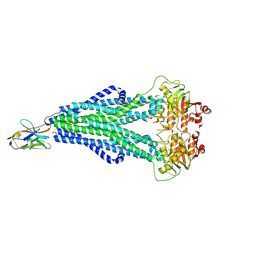 | | Enterococcus faecalis EfrCD in complex with a nanobody | | Descriptor: | ABC transporter ATP-binding protein, Nanobody | | Authors: | Ehrenbolger, K, Hutter, C.A.J, Meier, G, Seeger, M.A, Barandun, J. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Deep mutational scan of a drug efflux pump reveals its structure-function landscape.
Nat.Chem.Biol., 19, 2023
|
|
4UN0
 
 | | Crystal structure of the human CDK12-cyclinK complex | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Chaikuad, A, Goubin, S, Krojer, T, Sorrell, F.J, Nowak, R, Williams, E, Kopec, J, Mahajan, R.P, Burgess-Brown, N, Carpenter, E.P, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of the Cdk12/Cyck Complex with AMP-Pnp Reveal a Flexible C-Terminal Kinase Extension Important for ATP Binding.
Sci.Rep., 5, 2015
|
|
6MVQ
 
 | | HCV NS5B 1b N316 bound to Compound 31 | | Descriptor: | (4-{1-[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl]-1H-1,2,4-triazol-5-yl}-2-fluorophenyl)boronic acid, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
5O4Q
 
 | |
8SW3
 
 | | BG505 GT1.1 SOSIP in complex with NHP Fabs 12C11 and RM20A3 | | Descriptor: | 12C11 heavy chain variable region, 12C11 light chain variable region, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Torres, J.L, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Germline-targeting HIV vaccination induces neutralizing antibodies to the CD4 binding site.
Sci Immunol, 9, 2024
|
|
7OGS
 
 | |
5OKB
 
 | | High resolution structure of native Gan1D, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus | | Descriptor: | GLYCEROL, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.331 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
4UIB
 
 | | Crystal structure of 3p in complex with tafCPB | | Descriptor: | (2S)-6-AMINO-2-[[(1R)-1-(CYCLOHEXYLMETHYL)-2-OXO-2-[[(2S)-1,7,7-TRIMETHYLNORBORNAN-2-YL]AMINO]ETHYL]ARBAMOYLAMINO]HEXANOIC ACID, ACETATE ION, CARBOXYPEPTIDASE B, ... | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
8TZW
 
 | |
6O3L
 
 | |
8U08
 
 | |
8TZN
 
 | |
8U03
 
 | |
6NZJ
 
 | | Structural Analysis of a Nitrogenase Iron Protein from Methanosarcina acetivorans: Implications for CO2 Capture by a Surface-Exposed [Fe4S4] Cluster | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein, SULFATE ION | | Authors: | Rettberg, L.A, Kang, W, Stiebritz, M.T, Hiller, C.J, Lee, C.C, Liedtke, J, Ribbe, M.W, Hu, Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of a Nitrogenase Iron Protein from Methanosarcina acetivorans: Implications for CO 2 Capture by a Surface-Exposed [Fe 4 S 4 ] Cluster.
Mbio, 10, 2019
|
|
