4MAZ
 
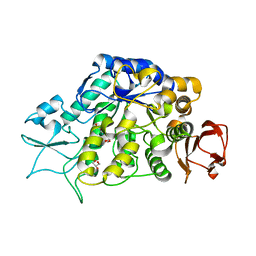 | | The Structure of MalL mutant enzyme V200S from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-18 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
6Z5E
 
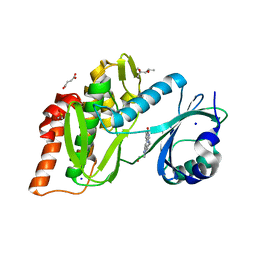 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004093 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 11-cyclopropyl-14-(2-hydroxyethyl)-8,11,14,18,19,22-hexazatetracyclo[13.5.2.12,6.018,21]tricosa-1(21),2(23),3,5,15(22),16,19-heptaen-7-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004093
To Be Published
|
|
8B6D
 
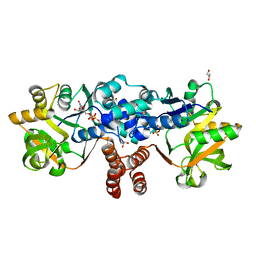 | |
6Z67
 
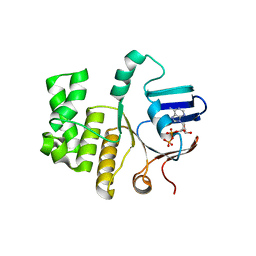 | | FtsE structure of Streptococcus pneumoniae in complex with AMPPNP at 2.4 A resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Alcorlo, M, Straume, D, Havarstein, L.S, Hermoso, j.A. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
6EDJ
 
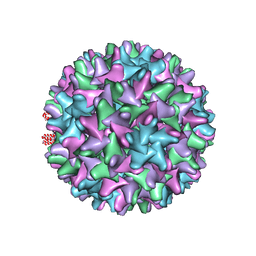 | | Cryo-EM structure of Woodchuck hepatitis virus capsid | | Descriptor: | External core antigen | | Authors: | Zhao, Z, Wang, J.C, Zlotnick, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Structural Differences between the Woodchuck Hepatitis Virus Core Protein in the Dimer and Capsid States Are Consistent with Entropic and Conformational Regulation of Assembly.
J.Virol., 93, 2019
|
|
8B31
 
 | |
6ZD8
 
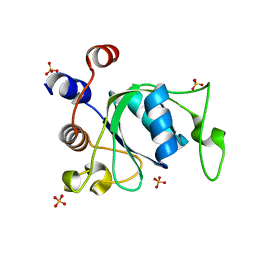 | | Crystal structure of YTHDC1 T379V mutant | | Descriptor: | SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
1U75
 
 | |
1KTC
 
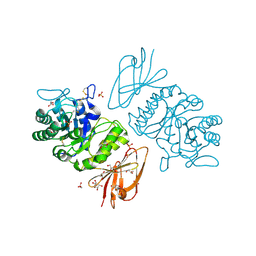 | | The Structure of alpha-N-Acetylgalactosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garman, S.C, Hannick, L, Zhu, A, Garboczi, D.N. | | Deposit date: | 2002-01-15 | | Release date: | 2002-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 1.9 A structure of alpha-N-acetylgalactosaminidase: molecular basis of glycosidase deficiency diseases.
Structure, 10, 2002
|
|
4LSZ
 
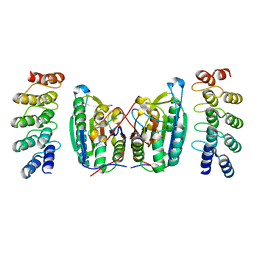 | | Caspase-7 in Complex with DARPin D7.18 | | Descriptor: | Caspase-7 subunit p10, Caspase-7 subunit p20, DARPin D7.18 | | Authors: | Fluetsch, A, Lukarska, M, Gruetter, M.G. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Combined inhibition of caspase 3 and caspase 7 by two highly selective DARPins slows down cellular demise.
Biochem.J., 461, 2014
|
|
3AE4
 
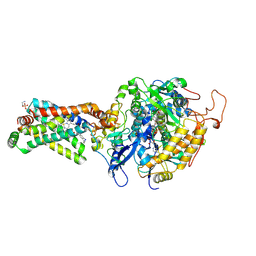 | | Crystal structure of porcine heart mitochondrial complex II bound with 2-Iodo-N-methyl-benzamide | | Descriptor: | 2-iodo-N-methylbenzamide, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with 2-Iodo-N-methyl-benzamide
To be Published
|
|
6ELF
 
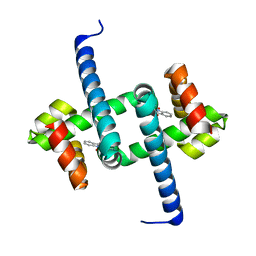 | | Tryptophan Repressor TrpR from E.coli variant M42F T44L T81I S88Y with Indole-3-acetic acid as ligand | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Trp operon repressor | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-09-28 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
1U0D
 
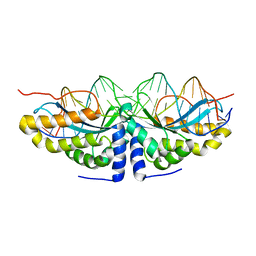 | | Y33H Mutant of Homing endonuclease I-CreI | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*CP*GP*C)-3', 5'-D(*GP*CP*GP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', DNA endonuclease I-CreI | | Authors: | Sussman, D, Chadsey, M, Fauce, S, Engel, A, Bruett, A, Monnat, R, Stoddard, B.L, Seligman, L.M. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Isolation and characterization of new homing endonuclease specificities at individual target site positions.
J.Mol.Biol., 342, 2004
|
|
4D81
 
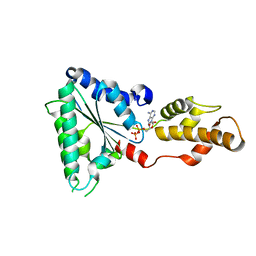 | | Metallosphera sedula Vps4 crystal structure | | Descriptor: | AAA ATPase, central domain protein, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Caillat, C, Macheboeuf, P, Wu, Y, McCarthy, A.A, Boeri-Erba, E, Effantin, G, Gottlinger, H.G, Weissenhorn, W, Renesto, P. | | Deposit date: | 2014-12-02 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Asymmetric Ring Structure of Vps4 Required for Escrt-III Disassembly.
Nat.Commun., 6, 2015
|
|
6ZEG
 
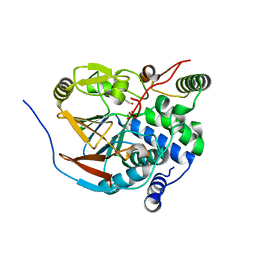 | | Structure of PP1-IRSp53 chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
1KHK
 
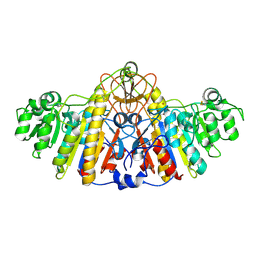 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) | | Descriptor: | Alkaline Phosphatase, MAGNESIUM ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
4V51
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with mRNA, tRNA and paromomycin | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Selmer, M, Dunham, C.M, Murphy, F.V, Weixlbaumer, A, Petry, S, Weir, J.R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2006-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 70S ribosome complexed with mRNA and tRNA.
Science, 313, 2006
|
|
2OPG
 
 | | The crystal structure of the 10th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein, SODIUM ION | | Authors: | Gileadi, C, Phillips, C, Elkins, J, Papagrigoriou, E, Ugochukwu, E, Gorrec, F, Savitsky, P, Umeano, C, Berridge, G, Gileadi, O, Salah, E, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the 10th PDZ domain of MPDZ
To be Published
|
|
4V5J
 
 | | Structure of the 70S ribosome bound to Release factor 2 and a substrate analog provides insights into catalysis of peptide release | | Descriptor: | 16S Ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jin, H, Kelley, A.C, Loakes, D, Ramakrishnan, V. | | Deposit date: | 2010-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
6ZJ1
 
 | | Structure of an inactive E404Q variant of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with tetrasaccharide N-glycan fragment and hexatungstotellurate(VI) TEW | | Descriptor: | 6-tungstotellurate(VI), Glycoprotein endo-alpha-1,2-mannosidase, MAGNESIUM ION, ... | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3NET
 
 | |
8BTM
 
 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GdmF | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional studies of geldanamycin amide synthase ShGdmF
To Be Published
|
|
6EQI
 
 | | Structure of PINK1 bound to ubiquitin | | Descriptor: | GLYCEROL, Nb696, Serine/threonine-protein kinase PINK1, ... | | Authors: | Schubert, A.F, Gladkova, C, Pardon, E, Wagstaff, J.L, Freund, S.M.V, Steyaert, J, Maslen, S, Komander, D. | | Deposit date: | 2017-10-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of PINK1 in complex with its substrate ubiquitin.
Nature, 552, 2017
|
|
6QOY
 
 | | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, 4-(2-azanylethyl)benzenesulfonic acid, CHLORIDE ION, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Kudryakova, I, Afoshin, A, Vasilyeva, N. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serine bacteriolytic protease L1 of Lysobacter sp. XL1 complexed with protease inhibitor AEBSF: features of interaction
Process Biochem, 2019
|
|
6ZJE
 
 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor Ap5A | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, CHLORIDE ION, GTP:AMP phosphotransferase AK3, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
