7AAQ
 
 | | sugar/H+ symporter STP10 in outward occluded conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ACETATE ION, ... | | Authors: | Bavnhoej, L, Paulsen, P.A, Pedersen, B.P. | | Deposit date: | 2020-09-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Molecular mechanism of sugar transport in plants unveiled by structures of glucose/H + symporter STP10.
Nat.Plants, 7, 2021
|
|
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
7A3A
 
 | |
7ACO
 
 | |
7ACM
 
 | |
6PGP
 
 | |
7Q2U
 
 | | The crystal structure of the HINT1 Q62A mutant. | | Descriptor: | CACODYLATE ION, HEXAETHYLENE GLYCOL, Histidine triad nucleotide-binding protein 1, ... | | Authors: | Dolot, R.M, Strom, A.M, Wagner, C.R. | | Deposit date: | 2021-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Dynamic Long-Range Interactions Influence Substrate Binding and Catalysis by Human Histidine Triad Nucleotide-Binding Proteins (HINTs), Key Regulators of Multiple Cellular Processes and Activators of Antiviral ProTides.
Biochemistry, 61, 2022
|
|
7A9K
 
 | |
6Y0N
 
 | | Arginine hydroxylase VioC in complex with Arg, 2OG and Fe under anaerobic environment using FT-SSX methods | | Descriptor: | 2-OXOGLUTARIC ACID, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, S.A, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Anaerobic fixed-target serial crystallography.
Iucrj, 7, 2020
|
|
7A9F
 
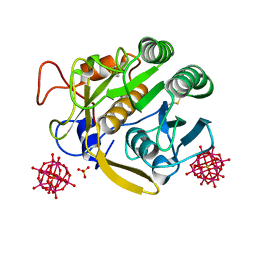 | |
7PGU
 
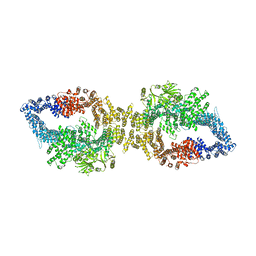 | | Autoinhibited structure of human neurofibromin isoform 2 stabilized by Zinc. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
6VAN
 
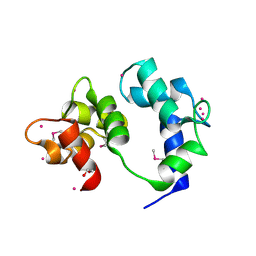 | | Crystal structure of caltubin from the great pond snail | | Descriptor: | 1,2-ETHANEDIOL, Caltubin, EF-hand, ... | | Authors: | Dong, A, Li, A, Zhang, Q, Barszczyk, A, Chern, Y.H, Arrowsmith, C.H, Edwards, A.M, Zhong, Z.P, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Cell-penetrating caltubin promotes neurite outgrowth and regrowth through calcium-dependent microtubule regulation
to be published
|
|
7PGT
 
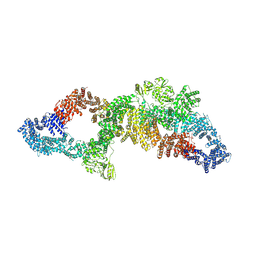 | | The structure of human neurofibromin isoform 2 in opened conformation. | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGS
 
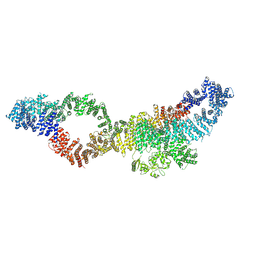 | | Consensus structure of human Neurofibromin isoform 2 | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
6PL5
 
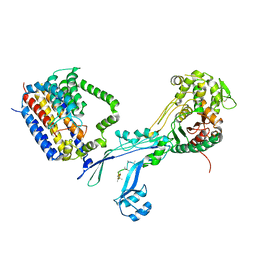 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, Unknown peptide | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
7ABR
 
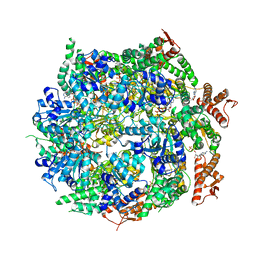 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | Authors: | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
7PGR
 
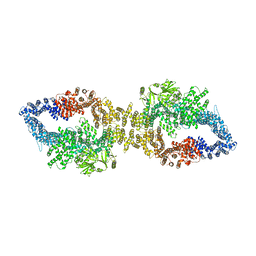 | | The structure of human neurofibromin isoform 2 in closed conformation | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7A6V
 
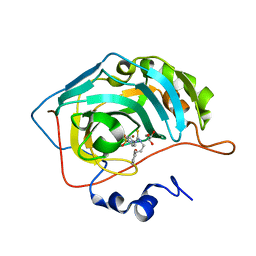 | |
7A9M
 
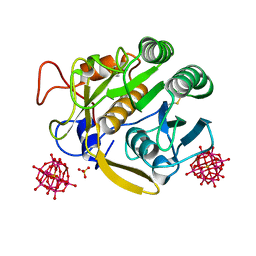 | |
7A8V
 
 | | Crystal structure of Polysaccharide monooxygenase from P.verruculosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Lytic polysaccharide monooxygenase, ... | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2020-08-31 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94961083 Å) | | Cite: | Crystal structure of Polysaccharide monooxygenase from P.verruculosum
To Be Published
|
|
8DLC
 
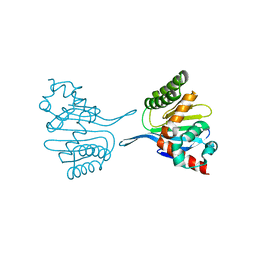 | | Crystal structure of chalcone-isomerase like protein from Vitis vinifera (VvCHIL) | | Descriptor: | Chalcone-flavonone isomerase family protein | | Authors: | Wolf Saxon, E, Moorman, C, Castro, A, Ruiz, A, Mallari, J.P, Burke, J.R. | | Deposit date: | 2022-07-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulatory ligand binding in plant chalcone isomerase-like (CHIL) proteins.
J.Biol.Chem., 299, 2023
|
|
6VC2
 
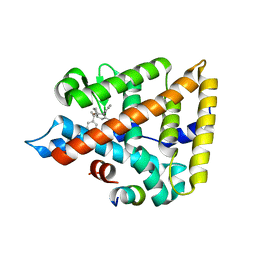 | | LRH-1 bound to SS-RJW100 and a fragment of the Tif2 Coactivator | | Descriptor: | (1S,3aS,6aS)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-ol, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Enantiomer-specific activities of an LRH-1 and SF-1 dual agonist.
Sci Rep, 10, 2020
|
|
8BBG
 
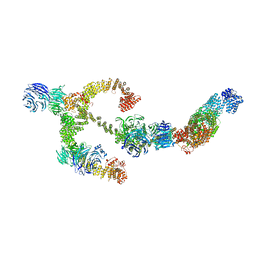 | | Structure of the IFT-A complex; anterograde IFT-A train model | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Intraflagellar transport protein 43 homolog, ... | | Authors: | Hesketh, S.J, Mukhopadhyay, A.G, Nakamura, D, Toropova, K, Roberts, A.J. | | Deposit date: | 2022-10-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | IFT-A structure reveals carriages for membrane protein transport into cilia.
Cell, 185, 2022
|
|
7A0O
 
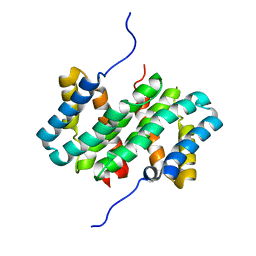 | | NMR structure of flagelliform spidroin (FlagSp) N-terminal domain from Trichonephila clavipes at pH 5.5 | | Descriptor: | Flagelliform spidroin variant 1 | | Authors: | Sarr, M, Kitoka, K, Walsh-White, K.-A, Kaldmae, M, Landreh, M, Rising, A, Johansson, J, Jaudzems, K, Kronqvist, N. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence.
J.Biol.Chem., 298, 2022
|
|
7PMF
 
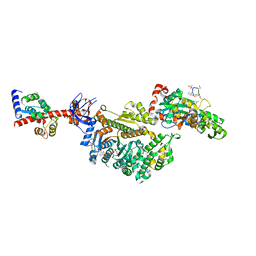 | | Cryo-EM structure of the actomyosin-V complex in the post-rigor transition state (AppNHp, central 1er, class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
