7TFE
 
 | | L. monocytogenes GS(12) - apo | | Descriptor: | Glutamine synthetase, MAGNESIUM ION | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7T1L
 
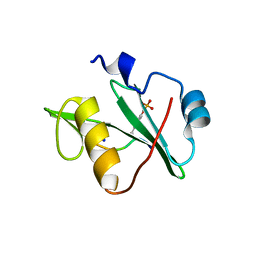 | | Crystal structure of a superbinder Fes SH2 domain (sFesS) in complex with a high affinity phosphopeptide | | Descriptor: | CHLORIDE ION, SODIUM ION, Synthetic phosphotyrosine-containing Ezrin-derived peptide, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7TEP
 
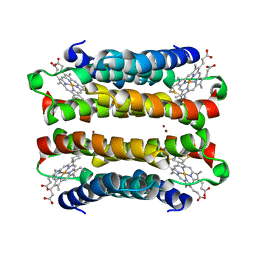 | |
2IXS
 
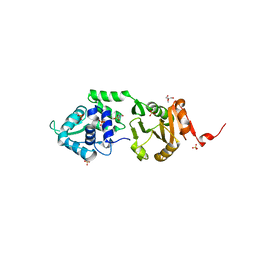 | | Structure of SdaI restriction endonuclease | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SDAI RESTRICTION ENDONUCLEASE, ... | | Authors: | Tamulaitiene, G, Jakubauskas, A, Urbanke, C, Huber, R, Grazulis, S, Siksnys, V. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Rare-Cutting Restriction Enzyme Sdai Reveals Unexpected Domain Architecture
Structure, 14, 2006
|
|
2IOA
 
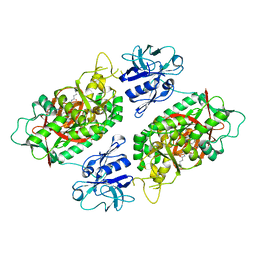 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ and ADP and phosphinate inhibitor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, D-GAMMA-GLUTAMYL-N-{[(R)-{4-[(4-AMINOBUTYL)AMINO]BUTYL}(PHOSPHONOOXY)PHOSPHORYL]METHYL}-D-ALANINAMIDE, ... | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
6J7W
 
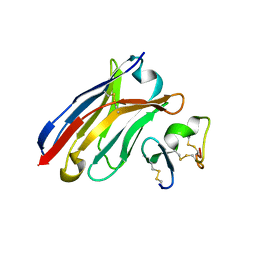 | | Crystal Structure of Human BCMA in complex with UniAb(TM) VH | | Descriptor: | Tumor necrosis factor receptor superfamily member 17, UniAb | | Authors: | Clarke, S.C, Ma, B, Trinklein, N.D, Schellenberger, U, Osborn, M, Ouisse, L, Boudreau, A, Davison, L, Harris, K.E, Ugamraj, H, Balasubramani, A, Dang, K, Jorgensen, B, Ogana, H, Pham, D, Pratap, P, Sankaran, P, Anegon, I, van Schooten, W, Bruggemann, M, Buelow, R, Force Aldred, S. | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Multispecific Antibody Development Platform Based on Human Heavy Chain Antibodies
Front Immunol, 9, 2018
|
|
7T1J
 
 | | Crystal structure of RUBISCO from Rhodospirillaceae bacterium BRH_c57 | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Pereira, J.H, Liu, A.K, Shih, P.M, Adams, P.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural plasticity enables evolution and innovation of RuBisCO assemblies.
Sci Adv, 8, 2022
|
|
1T95
 
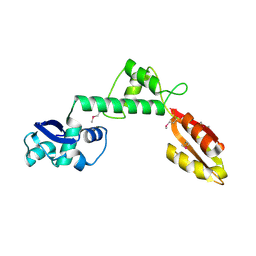 | |
7SQK
 
 | | Cryo-EM structure of the human augmin complex | | Descriptor: | HAUS augmin-like complex subunit 1, HAUS augmin-like complex subunit 2, HAUS augmin-like complex subunit 3, ... | | Authors: | Gabel, C.A, Chang, L. | | Deposit date: | 2021-11-05 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Molecular architecture of the augmin complex.
Nat Commun, 13, 2022
|
|
4S20
 
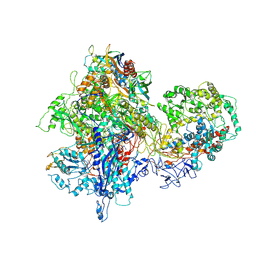 | | Structural basis for transcription reactivation by RapA | | Descriptor: | 5'-D(P*AP*CP*GP*AP*CP*TP*GP*AP*GP*CP*CP*GP*AP*TP*G)-3', 5'-R(P*AP*UP*CP*GP*GP*CP*UP*CP*A)-3', DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Liu, B, Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-01-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Structural basis for transcription reactivation by RapA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7T2V
 
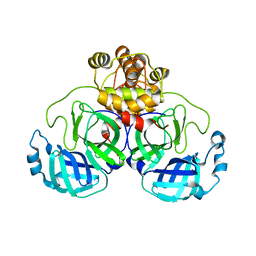 | |
7T2U
 
 | |
7STO
 
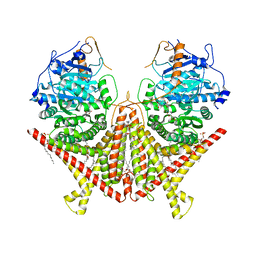 | | Chitin Synthase 2 from Candida albicans bound to polyoxin D | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-{(2R,3R,4S,5R)-5-[(S)-{[(2S,3S,4S)-2-amino-5-(carbamoyloxy)-3,4-dihydroxypentanoyl]amino}(carboxy)methyl]-3,4-dihydroxyoxolan-2-yl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid (non-preferred name), Chitin synthase | | Authors: | Ren, Z, Chhetri, A, Lee, S, Yokoyama, K. | | Deposit date: | 2021-11-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for inhibition and regulation of a chitin synthase from Candida albicans.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7STM
 
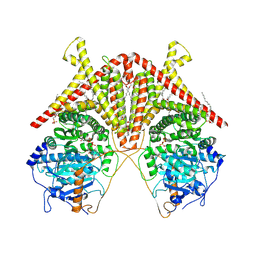 | | Chitin Synthase 2 from Candida albicans bound to UDP-GlcNAc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Chitin synthase, MAGNESIUM ION, ... | | Authors: | Ren, Z, Chhetri, A, Lee, S, Yokoyama, K. | | Deposit date: | 2021-11-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis for inhibition and regulation of a chitin synthase from Candida albicans.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7STN
 
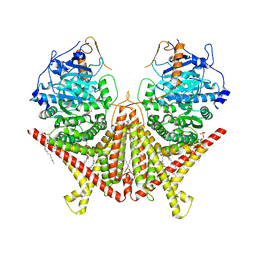 | | Chitin Synthase 2 from Candida albicans bound to Nikkomycin Z | | Descriptor: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), 1,2-Distearoyl-sn-glycerophosphoethanolamine, Chitin synthase | | Authors: | Ren, Z, Chhetri, A, Lee, S, Yokoyama, K. | | Deposit date: | 2021-11-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for inhibition and regulation of a chitin synthase from Candida albicans.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2I26
 
 | |
4QQG
 
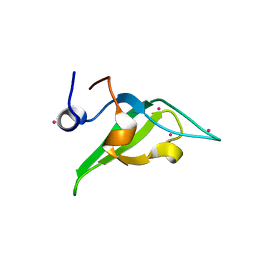 | | Crystal structure of an N-terminal HTATIP fragment | | Descriptor: | Histone acetyltransferase KAT5, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Wernimont, A.K, Dobrovetsky, E, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and histone binding studies of the chromo barrel domain of TIP60.
FEBS Lett., 592, 2018
|
|
3E4U
 
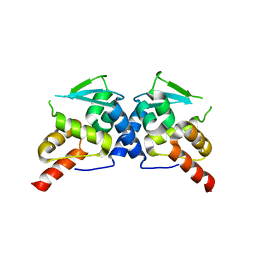 | | Crystal Structure of the Wild-Type Human BCL6 BTB/POZ Domain | | Descriptor: | B-cell lymphoma 6 protein | | Authors: | Stead, M.A, Rosbrook, G.O, Hadden, J.M, Trinh, C.H, Carr, S.B, Wright, S.C. | | Deposit date: | 2008-08-12 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the wild-type human BCL6 POZ domain.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
6VRA
 
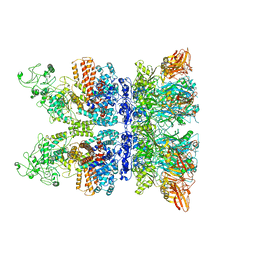 | | Anthrax octamer prechannel bound to full-length edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Zhou, K, Hardenbrook, N.J, Liu, S, Cui, Y.X, Krantz, B.A, Zhou, Z.H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic Structures of Anthrax Prechannel Bound with Full-Length Lethal and Edema Factors.
Structure, 28, 2020
|
|
1TKN
 
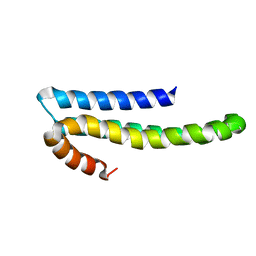 | | Solution structure of CAPPD*, an independently folded extracellular domain of human Amyloid-beta Precursor Protein | | Descriptor: | Amyloid beta A4 protein | | Authors: | Dulubova, I, Ho, A, Huryeva, I, Sudhof, T.C, Rizo, J. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of an independently folded extracellular domain of human amyloid-beta precursor protein.
Biochemistry, 43, 2004
|
|
5K5G
 
 | | Structure of human islet amyloid polypeptide in complex with an engineered binding protein | | Descriptor: | HI18, Islet amyloid polypeptide | | Authors: | Mirecka, E.A, Feuerstein, S, Gremer, L, Schroeder, G.F, Stoldt, M, Willbold, D, Hoyer, W. | | Deposit date: | 2016-05-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | beta-Hairpin of Islet Amyloid Polypeptide Bound to an Aggregation Inhibitor.
Sci Rep, 6, 2016
|
|
1JY9
 
 | | MINIMIZED AVERAGE STRUCTURE OF DP-TT2 | | Descriptor: | DP-TT2 | | Authors: | Stanger, H.E, Syud, F.A, Espinosa, J.F, Giriat, I, Muir, T, Gellman, S.H. | | Deposit date: | 2001-09-11 | | Release date: | 2001-09-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Length-dependent stability and strand length limits in antiparallel beta -sheet secondary structure.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4RA7
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-2-hydroxy-1-{[(2-hydroxynaphthalen-1-yl)carbonyl]amino}ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin
To be Published
|
|
1SW2
 
 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with glycine betaine | | Descriptor: | TRIMETHYL GLYCINE, osmoprotection protein (proX) | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
4IDT
 
 | | Crystal Structure of NIK with 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine (T28) | | Descriptor: | 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Liu, J, Sudom, A, Wang, Z. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibiting NF-KB-inducing kinase (NIK): Discovery, structure-based design, synthesis, structure activity relationship, and co-crystal structures
Bioorg.Med.Chem.Lett., 23, 2013
|
|
