2I7G
 
 | | Crystal Structure of Monooxygenase from Agrobacterium tumefaciens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Monooxygenase, SULFATE ION | | Authors: | Kim, Y, Xu, X, Zheng, H, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of Monooxygenase from Agrobacterium tumefaciens
To be Published, 2006
|
|
7T4L
 
 | | Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with extended alpha-C helix in chain B | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
6CKY
 
 | | Crystal structure of UcmS2 | | Descriptor: | Glyoxalase | | Authors: | Chang, C.Y, Chang, C, Annaval, T, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of UcmS2
To Be Published
|
|
1T0S
 
 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase with 4-bromophenol bound | | Descriptor: | 4-BROMOPHENOL, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Sazinsky, M.H, Bard, J, Di Donato, A, Lippard, S.J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase from Pseudomonas stutzeri OX1: INSIGHT INTO THE SUBSTRATE SPECIFICITY, SUBSTRATE CHANNELING, AND ACTIVE SITE TUNING OF MULTICOMPONENT MONOOXYGENASES.
J.Biol.Chem., 279, 2004
|
|
6VP7
 
 | |
3ZOT
 
 | | Structure of E.coli rhomboid protease GlpG in complex with monobactam L29 (data set 2) | | Descriptor: | CHLORIDE ION, RHOMBOID PROTEASE GLPG, nonyl beta-D-glucopyranoside, ... | | Authors: | Vinothkumar, K.R, Pierrat, O.A, Large, J.M, Freeman, M. | | Deposit date: | 2013-02-24 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structure of Rhomboid Protease in Complex with Beta-Lactam Inhibitors Defines the S2' Cavity.
Structure, 21, 2013
|
|
2I0F
 
 | | Lumazine synthase RibH1 from Brucella abortus (Gene BruAb1_0785, Swiss-Prot entry Q57DY1) | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
2I0Z
 
 | | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-utilizing dehydrogenases | | Authors: | Minasov, G, Shuvalova, L, Vorontsov, I.I, Kiryukhina, O, Abdullah, J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases
To be Published
|
|
1T4D
 
 | | Crystal structure of Escherichia coli aspartate beta-semialdehyde dehydrogenase (EcASADH), at 1.95 Angstrom resolution | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Nichols, C.E, Dhaliwal, B, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-04-29 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution Structures Reveal Details of Domain Closure and "Half-of-sites-reactivity" in Escherichia coli Aspartate beta-Semialdehyde Dehydrogenase.
J.Mol.Biol., 341, 2004
|
|
6W0A
 
 | | Open-gate KcsA soaked in 1 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.237 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
5JZY
 
 | | Thrombin in complex with (S)-1-((R)-2-amino-3-cyclohexylpropanoyl)-N-(4-carbamimidoylbenzyl)pyrrolidine-2-carboxamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-cyclohexyl-D-alanyl-N-[(4-carbamimidoylphenyl)methyl]-L-prolinamide, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2016-05-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Strategies for Late-Stage Optimization: Profiling Thermodynamics by Preorganization and Salt Bridge Shielding.
J.Med.Chem., 62, 2019
|
|
6W0I
 
 | | Closed-gate KcsA soaked in 10mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W43
 
 | | APE1 AP-endonuclease product complex R237C | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6W0Q
 
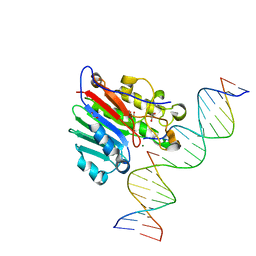 | | APE1 endonuclease product complex D148E | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-02 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
4R85
 
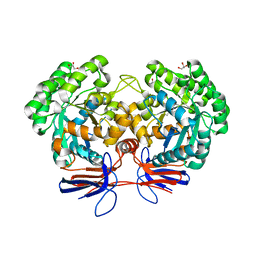 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-methylcytosine | | Descriptor: | 5-methylcytosine, Cytosine deaminase, FE (II) ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-methylcytosine
To be Published
|
|
3ECR
 
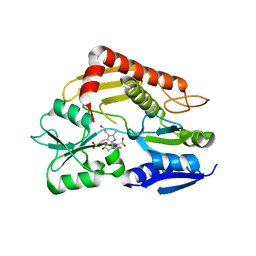 | | Structure of human porphobilinogen deaminase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Song, G, Li, Y, Cheng, C, Zhao, Y, Gao, A, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural insight into acute intermittent porphyria.
Faseb J., 23, 2009
|
|
3EDI
 
 | |
2VWJ
 
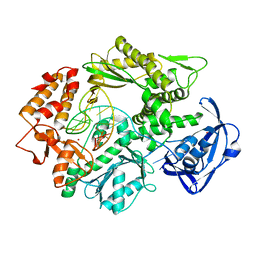 | | Uracil Recognition in Archaeal DNA Polymerases Captured by X-ray Crystallography. | | Descriptor: | 5'-D(*AP*AP*UP*GP*GP*AP*GP*AP*CP*GP *GP*CP*TP*TP*TP*TP*GP*CP*CP*GP*TP*GP*TP*C)-3', DNA POLYMERASE, POTASSIUM ION | | Authors: | Firbank, S.J, Wardle, J, Heslop, P, Lewis, R.J, Connolly, B.A. | | Deposit date: | 2008-06-25 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Uracil Recognition in Archaeal DNA Polymerases Captured by X-Ray Crystallography.
J.Mol.Biol., 381, 2008
|
|
3X2H
 
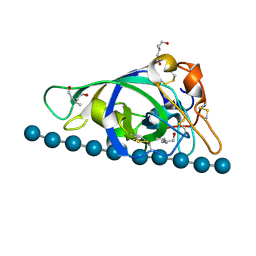 | | X-ray structure of PcCel45A N92D with cellopentaose at 95K. | | Descriptor: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
2I8A
 
 | |
7TDM
 
 | |
1TAF
 
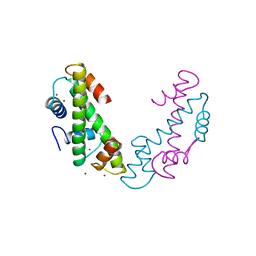 | | DROSOPHILA TBP ASSOCIATED FACTORS DTAFII42/DTAFII62 HETEROTETRAMER | | Descriptor: | TFIID TBP ASSOCIATED FACTOR 42, TFIID TBP ASSOCIATED FACTOR 62, ZINC ION | | Authors: | Xie, X, Kokubo, T, Cohen, S.L, Mirza, U.A, Hoffmann, A, Chait, B.T, Roeder, R.G, Nakatani, Y, Burley, S.K. | | Deposit date: | 1996-06-01 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural similarity between TAFs and the heterotetrameric core of the histone octamer.
Nature, 380, 1996
|
|
4QQE
 
 | | Crystal structure of WDR5, WD repeat domain 5 in complex with compound SGC-DS-MT-0345 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-[2-(4-methylpiperazin-1-yl)-5-(quinolin-3-yl)phenyl]-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Wernimont, A, Smil, D, Getlik, M, Senisterra, G, Poda, G, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of WDR5, WD repeat domain 5 in complex with compound SGC-DS-MT-0345
To be Published
|
|
7TDN
 
 | |
5K7W
 
 | |
