4FQR
 
 | |
1BGK
 
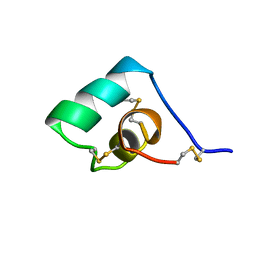 | | SEA ANEMONE TOXIN (BGK) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL, NMR, 15 STRUCTURES | | Descriptor: | BGK | | Authors: | Dauplais, M, Lecoq, A, Song, J, Cotton, J, Jamin, N, Gilquin, B, Roumestand, C, Vita, C, Harvey, A, Menez, A. | | Deposit date: | 1996-05-08 | | Release date: | 1997-01-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures.
J.Biol.Chem., 272, 1997
|
|
1BHA
 
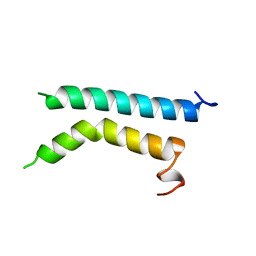 | | THREE-DIMENSIONAL STRUCTURE OF (1-71) BACTERIOOPSIN SOLUBILIZED IN METHANOL-CHLOROFORM AND SDS MICELLES DETERMINED BY 15N-1H HETERONUCLEAR NMR SPECTROSCOPY | | Descriptor: | BACTERIORHODOPSIN | | Authors: | Pervushin, K.V, Orekhov, V.Y, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | Deposit date: | 1993-10-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
6ETU
 
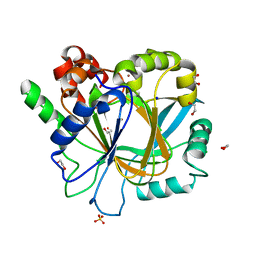 | | Crystal structure of KDM4D with tetrazolhydrazide compound 7 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
4F65
 
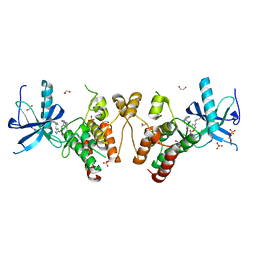 | | Crystal structure of Human Fibroblast Growth Factor Receptor 1 Kinase domain in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N~2~-[(3-methyl-1,2-oxazol-5-yl)methyl]-N~4~-[3-(2-phenylethyl)-1H-pyrazol-5-yl]pyrimidine-2,4-diamine, Fibroblast growth factor receptor 1, ... | | Authors: | Norman, R.A, Breed, J, Ogg, D. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Protein-Ligand Crystal Structures Can Guide the Design of Selective Inhibitors of the FGFR Tyrosine Kinase.
J.Med.Chem., 55, 2012
|
|
4YW6
 
 | | Structural Insight into Divalent Galactoside Binding to Pseudomonas aeruginosa lectin LecA | | Descriptor: | CALCIUM ION, N-[(2S)-6-amino-1-oxo-1-(pyrrolidin-1-yl)hexan-2-yl]-4-(beta-D-galactopyranosyloxy)benzamide, PA-I galactophilic lectin | | Authors: | Visini, R, Jin, X, Michaud, G, Bergmann, M, Gillon, E, Imberty, A, Stocker, A, Darbre, T, Pieters, R, Reymond, J.-L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insight into Multivalent Galactoside Binding to Pseudomonas aeruginosa Lectin LecA.
Acs Chem.Biol., 10, 2015
|
|
6F8O
 
 | | AKR1B1 at 3.45 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
4YO9
 
 | | HKU4 3CLpro unbound structure | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like proteinase, ACETATE ION, ... | | Authors: | St John, S.E, Mesecar, A. | | Deposit date: | 2015-03-11 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS).
Bioorg.Med.Chem., 23, 2015
|
|
6F90
 
 | | Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Alpha-1,2-mannosidase, putative, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6F9P
 
 | | Crystal structure of the Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO5 with Re(II) | | Descriptor: | CHLORIDE ION, GLYCEROL, L-lysine 4-hydroxylase, ... | | Authors: | Isabet, T, Stura, E, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
4YWH
 
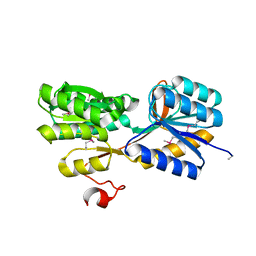 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM ACTINOBACILLUS SUCCINOGENES 130Z (Asuc_0499, TARGET EFI-511068) WITH BOUND D-XYLOSE | | Descriptor: | ABC TRANSPORTER SOLUTE BINDING PROTEIN, beta-D-xylopyranose | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM ACTINOBACILLUS SUCCINOGENES 130Z (Asuc_0499, TARGET EFI-511068) WITH BOUND D-XYLOSE
To be published
|
|
7SHJ
 
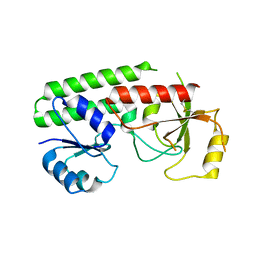 | |
3M5A
 
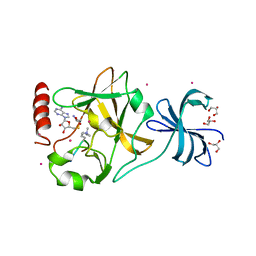 | | SET7/9 Y245A in complex with TAF10-K189me3 peptide and AdoHcy | | Descriptor: | COBALT (II) ION, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
4Y3R
 
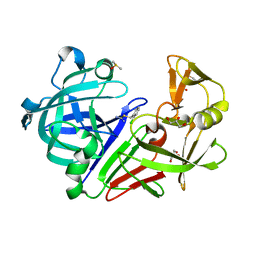 | | Endothiapepsin in complex with fragment 306 | | Descriptor: | 2-chlorobenzyl carbamimidothioate, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, X, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
5GZU
 
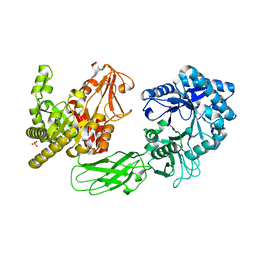 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | Chitinase, PHOSPHATE ION | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
6MGX
 
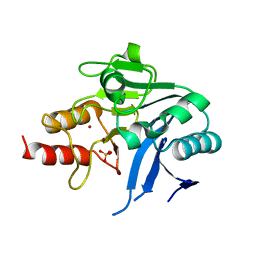 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 6 Klebsiella pneumoniae | | Descriptor: | Metallo-beta-lactamase, SULFATE ION, ZINC ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 6 Klebsiella pneumoniae
To Be Published
|
|
4YQY
 
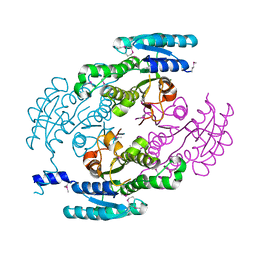 | | Crystal Structure of a putative Dehydrogenase from Sulfitobacter sp. (COG1028) (TARGET EFI-513936) in its APO form | | Descriptor: | MAGNESIUM ION, Putative Dehydrogenase | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-13 | | Release date: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Crystal Structure of a putative Dehydrogenase from Sulfitobacter sp. (COG1028, TARGET EFI-513936) in its APO form
To be published
|
|
6BF8
 
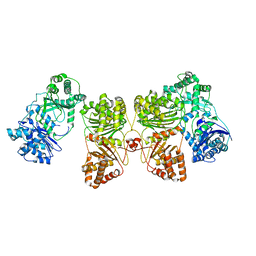 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
5YBG
 
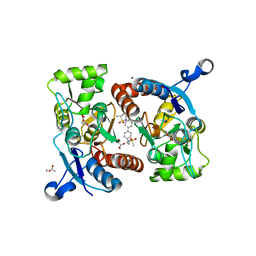 | | Crystal structure of the GluA2o LBD in complex with glutamate and LY451395 | | Descriptor: | ACETATE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | HBT1, a Novel AMPA Receptor Potentiator with Lower Agonistic Effect, Avoided Bell-Shaped Response in In Vitro BDNF Production.
J. Pharmacol. Exp. Ther., 364, 2018
|
|
6EXW
 
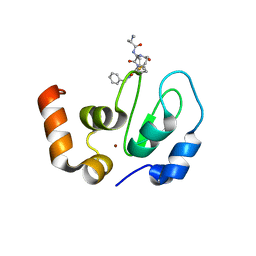 | | Crystal structure of cIAP1-BIR3 in complex with a covalently bound SM | | Descriptor: | (3~{S},6~{S},7~{R},9~{a}~{S})-6-[[(2~{S})-2-(methylamino)propanoyl]amino]-5-oxidanylidene-~{N}-(phenylmethyl)-7-[(propanoylamino)methyl]-3,6,7,8,9,9~{a}-hexahydropyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Corti, A, Cossu, F, Milani, M, Mastrangelo, E. | | Deposit date: | 2017-11-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design and molecular profiling of Smac-mimetics selective for cellular IAPs.
FEBS J., 285, 2018
|
|
3M7Q
 
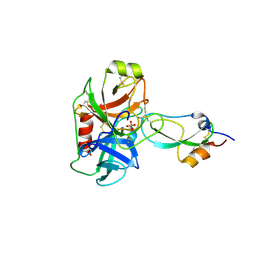 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean sea anemone stichodactyla helianthus in complex with bovine pancreatic trypsin | | Descriptor: | Cationic trypsin, Kunitz-type proteinase inhibitor SHPI-1, PHOSPHATE ION | | Authors: | Garcia-Fernandez, R, Redecke, L, Pons, T, Perbandt, M, Gil, D, Talavera, A, Gonzalez, Y, de los angeles Chavez, M, Betzel, C. | | Deposit date: | 2010-03-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into serine protease inhibition by a marine invertebrate BPTI Kunitz-type inhibitor.
J.Struct.Biol., 180, 2012
|
|
7ZE0
 
 | | Solution structure of the PulM C-terminal domain from Klebsiella oxytoca | | Descriptor: | Type II secretion system protein M | | Authors: | Lopez-Castilla, A, Bardiaux, B, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2022-03-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
7N3C
 
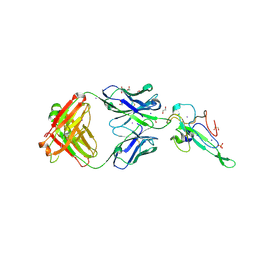 | | Crystal Structure of Human Fab S24-202 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Nucleoprotein, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7N3D
 
 | | Crystal Structure of Human Fab S24-1564 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nucleoprotein, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
6Y7J
 
 | | Structure of the BRD9 bromodomain and compound 15 | | Descriptor: | 1-[8-(2,5-dimethoxyphenyl)pyrrolo[1,2-a]pyrimidin-6-yl]ethanone, ACETATE ION, Bromodomain-containing protein 9, ... | | Authors: | Diaz-Saez, L, Krojer, T, Picaud, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M. | | Deposit date: | 2020-03-01 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the BRD9 bromodomain
To Be Published
|
|
