5LW1
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_232_11_D12 in complex JNK1a1 and JIP1 peptide | | Descriptor: | ADENOSINE, C-Jun-amino-terminal kinase-interacting protein 1, DD_232_11_D12, ... | | Authors: | Wu, Y, Batyuk, A, Mittl, P.R, Honegger, A, Plueckthun, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Selective Inhibition of c-Jun N-Terminal Kinase 1 Determined by Rigid DARPin-DARPin Fusions.
J.Mol.Biol., 430, 2018
|
|
7BH5
 
 | | XFEL structure of the ertapenem-derived CTX-M-15 acylenzyme after mixing for 2 sec using a piezoelectric injector (PolyPico) | | Descriptor: | (2~{S},3~{R},4~{R})-3-[5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-4-methyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BHM
 
 | |
7SES
 
 | | PRMT5/MEP50 with compound 29 bound | | Descriptor: | (2P)-2-{4-[4-(aminomethyl)-1-oxo-1,2-dihydrophthalazin-6-yl]-1-methyl-1H-pyrazol-5-yl}naphthalene-1-carbonitrile, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Smith, C.R, Kulyk, S, Marx, M.A. | | Deposit date: | 2021-10-01 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
6WIS
 
 | | Copper resistance protein copG- Form 1 | | Descriptor: | ACETATE ION, COPPER (II) ION, DUF411 domain-containing protein, ... | | Authors: | Hausrath, A.C, Ly, A, McEvoy, M.M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-06-24 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The bacterial copper resistance protein CopG contains a cysteine-bridged tetranuclear copper cluster.
J.Biol.Chem., 295, 2020
|
|
7BH7
 
 | | Room temperature, serial X-ray structure of the ertapenem-derived acylenzyme of CTX-M-15 (10 min soak) collected on fixed target chips at Diamond Light Source I24 | | Descriptor: | (2~{S},3~{R},4~{R})-3-[5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-4-methyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
5N4T
 
 | | VIM-2 metallo-beta-lactamase in complex with ((S)-3-mercapto-2-methylpropanoyl)-L-tryptophan (Compound 4) | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[[(2~{S})-2-methyl-3-sulfanyl-propanoyl]amino]propanoic acid, BENZOIC ACID, Beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
6ND6
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with erythromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.85A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
7RTP
 
 | |
6YZG
 
 | |
5N8D
 
 | |
7BII
 
 | | Crystal structure of Nematocida HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Grabarczyk, D.B, Petrova, O.A, Meinhart, A, Kessler, D, Clausen, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.037 Å) | | Cite: | HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain.
Nat.Chem.Biol., 17, 2021
|
|
8EEB
 
 | | Cryo-EM structure of human ABCA7 in Digitonin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phospholipid-transporting ATPase ABCA7, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
6MZJ
 
 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 2 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
6Z2A
 
 | |
8WDU
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
5N8C
 
 | | Crystal structure of Pseudomonas aeruginosa LpxC complexed with inhibitor | | Descriptor: | (2~{S})-3-azanyl-2-[[(1~{R})-5-[2-[4-[[2-(hydroxymethyl)imidazol-1-yl]methyl]phenyl]ethynyl]-2,3-dihydro-1~{H}-inden-1-yl]amino]-3-methyl-~{N}-oxidanyl-butanamide, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Cross, J.B, Ryan, M.D, Zhang, J, Cheng, R.K, Wood, M, Andersen, O.A, Brooks, M, Kwong, J, Barker, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based discovery of LpxC inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8GFN
 
 | | Room temperature X-ray structure of truncated SARS-CoV-2 main protease C145A mutant, residues 1-304, in complex with BBH1 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-(1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Coates, L. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors.
J.Biol.Chem., 299, 2023
|
|
6PZY
 
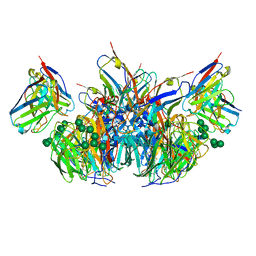 | | CryoEM derived model of NA-73 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-73 fragment antibody heavy chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6XV4
 
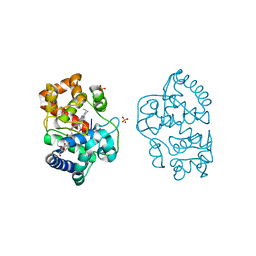 | | Neutron structure of ferric ascorbate peroxidase-ascorbate complex | | Descriptor: | ASCORBIC ACID, Ascorbate peroxidase, POTASSIUM ION, ... | | Authors: | Kwon, H, Basran, J, Devos, J.M, Schrader, T.E, Ostermann, A, Blakeley, M.P, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2020-01-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the protons in a metalloenzyme electron proton transfer pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XX0
 
 | | Crystal structure of NEMO in complex with Ubv-LIN | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b, ... | | Authors: | Akutsu, M, Skenderovic, A, Garcia-Pardo, J, Maculins, T, Dikic, I. | | Deposit date: | 2020-01-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
7OGJ
 
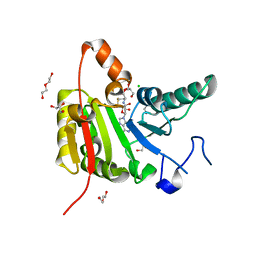 | |
6TPN
 
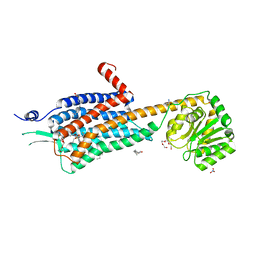 | | Crystal structure of the Orexin-2 receptor in complex with HTL6641 at 2.61 A resolution | | Descriptor: | 2-(5,6-dimethoxypyridin-3-yl)-1,1-bis(oxidanylidene)-4-[[2,4,6-tris(fluoranyl)phenyl]methyl]pyrido[2,3-e][1,2,4]thiadiazin-3-one, NITRATE ION, OLEIC ACID, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
5NC9
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (2S)-2,6-diamino-N-hydroxyhexanamide | | Descriptor: | (2~{S})-2,6-bis(azanyl)-~{N}-oxidanyl-hexanamide, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NCN
 
 | | Crystal structure Dbf2(NTR)-Mob1 complex | | Descriptor: | CHLORIDE ION, Cell cycle protein kinase DBF2, DBF2 kinase activator protein MOB1, ... | | Authors: | Gogl, G, Remenyi, A, Parker, B, Weiss, E. | | Deposit date: | 2017-03-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Ndr/Lats Kinases Bind Specific Mob-Family Coactivators through a Conserved and Modular Interface.
Biochemistry, 2020
|
|
