8T7E
 
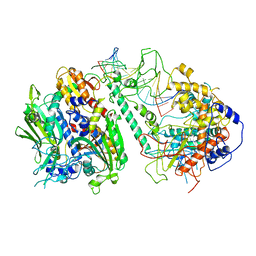 | | Cryo-EM structure of the Backtracking Initiation Complex (VII) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-06-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
7NHE
 
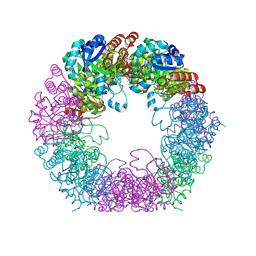 | | Crystal structure of Arabidopsis thaliana Pdx1K166R-I333 complex | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, [(~{E},4~{S})-4-azanyl-3-oxidanylidene-pent-1-enyl] dihydrogen phosphate | | Authors: | Rodrigues, M.J, Zhang, Y, Bolton, R, Evans, G, Giri, N, Royant, A, Begley, T, Ealick, S.E, Tews, I. | | Deposit date: | 2021-02-10 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Trapping and structural characterisation of a covalent intermediate in vitamin B 6 biosynthesis catalysed by the Pdx1 PLP synthase.
Rsc Chem Biol, 3, 2022
|
|
6MCD
 
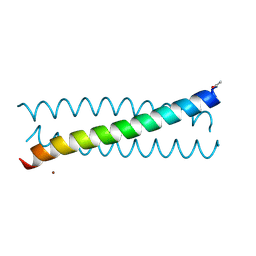 | | Crystal Structure of tris-thiolate Pb(II) complex with adjacent water in a de novo Three Stranded Coiled Coil Peptide | | Descriptor: | LEAD (II) ION, Pb(II)(GRAND Coil Ser L12CL16A)-, ZINC ION | | Authors: | Tolbert, A.E, Ruckthong, L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2018-08-31 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Heteromeric three-stranded coiled coils designed using a Pb(II)(Cys)3template mediated strategy.
Nat.Chem., 12, 2020
|
|
5M1Q
 
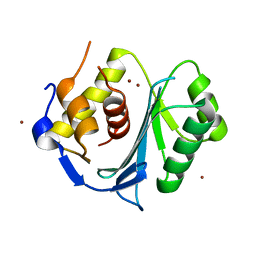 | | Crystal structure of the large terminase nuclease from thermophilic phage G20c with bound Zinc | | Descriptor: | Phage terminase large subunit, ZINC ION | | Authors: | Xu, R.G, Jenkins, H.T, Chechik, M, Blagova, E.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Viral genome packaging terminase cleaves DNA using the canonical RuvC-like two-metal catalysis mechanism.
Nucleic Acids Res., 45, 2017
|
|
5M30
 
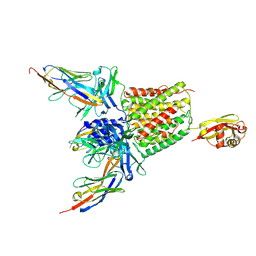 | | Structure of TssK from T6SS EAEC in complex with nanobody nb18 | | Descriptor: | Anti-vesicular stomatitis virus N VHH, Type VI secretion protein | | Authors: | Nguyen, V.S, Cambillau, C, Spinelli, C, Desmyter, A, Legrand, P, Cascales, E. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-21 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
7PK6
 
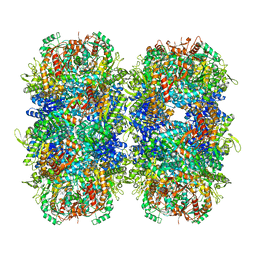 | | Providencia stuartii Arginine decarboxylase (Adc), stack structure | | Descriptor: | Biodegradative arginine decarboxylase | | Authors: | Jessop, M, Desfosses, A, Bacia-Verloop, M, Gutsche, I. | | Deposit date: | 2021-08-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Structural and biochemical characterisation of the Providencia stuartii arginine decarboxylase shows distinct polymerisation and regulation.
Commun Biol, 5, 2022
|
|
5LP6
 
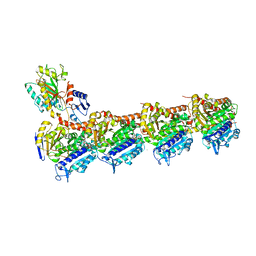 | | Crystal structure of Tubulin-Stathmin-TTL-Thiocolchicine Complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Marangon, J, Christodoulou, M, Casagrande, F, Tiana, G, Dalla Via, L, Aliverti, A, Passarella, D, Cappelletti, G, Ricagno, S. | | Deposit date: | 2016-08-11 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Tools for the rational design of bivalent microtubule-targeting drugs.
Biochem. Biophys. Res. Commun., 479, 2016
|
|
8OK2
 
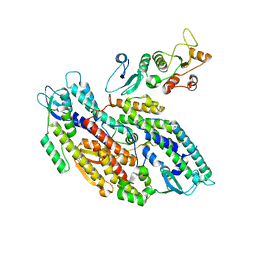 | | Bipartite interaction of TOPBP1 with the GINS complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, DNA replication complex GINS protein PSF3, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2023-03-26 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | TopBP1 utilises a bipartite GINS binding mode to support genome replication.
Nat Commun, 15, 2024
|
|
7Q1G
 
 | |
4XR8
 
 | | Crystal structure of the HPV16 E6/E6AP/p53 ternary complex at 2.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Martinez-Zapien, D, Ruiz, F.X, Mitschler, A, Podjarny, A, Trave, G, Zanier, K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.
Nature, 529, 2016
|
|
8T1N
 
 | | Micro-ED Structure of a Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Cascio, D, Sawaya, M.R, Cannon, K.A, Rodriguez, J.A, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7ACZ
 
 | |
7ACY
 
 | |
7ACV
 
 | |
7ACW
 
 | |
5LUJ
 
 | |
8TH2
 
 | | Structure of the isoflavene-forming dirigent protein PsPTS2 | | Descriptor: | Dirigent protein | | Authors: | Smith, C.A, Meng, Q, Lewis, N.G, Davin, L.B. | | Deposit date: | 2023-07-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dirigent isoflavene-forming PsPTS2: 3D structure, stereochemical, and kinetic characterization comparison with pterocarpan-forming PsPTS1 homolog in pea.
J.Biol.Chem., 300, 2024
|
|
8OR7
 
 | | Structure of a far-red induced allophycocyanin from Chroococcidiopsis thermalis sp. PCC 7203 | | Descriptor: | Allophycocyanin beta subunit apoprotein, PHYCOCYANOBILIN, POTASSIUM ION, ... | | Authors: | Zhou, L.J, Hoeppner, A, Wang, Y.Q, Zhao, K.H. | | Deposit date: | 2023-04-13 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic and biochemical analyses of a far-red allophycocyanin to address the mechanism of the super-red-shift.
Photosynth.Res., 2024
|
|
5LIZ
 
 | | The structure of Nt.BspD6I nicking endonuclease with all cysteines mutated by serine residues at 0.19 nm resolution . | | Descriptor: | Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Artyukh, R.I, Perevyazova, T.A, Yunusova, A.K, Popov, A.N, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural features of Cysteine residues mutation of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
6LTG
 
 | | Crystal structure of the Fab fragment of murine monoclonal antibody OHV-3 against Human herpesvirus 6B | | Descriptor: | MAGNESIUM ION, antibody Fab fragment H-chain, antibody Fab fragment L-chain | | Authors: | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
6RFF
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 7 | | Descriptor: | (~{E})-~{N}-(5-bromanyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-nitro-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-14 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
6RP9
 
 | | Crystal structure of the T-cell receptor NYE_S3 bound to HLA A2*01-SLLMWITQV | | Descriptor: | Beta-2-microglobulin, Cancer/testis antigen 1, HLA class I histocompatibility antigen, ... | | Authors: | Coles, C.H, Mulvaney, R, Malla, S, Lloyd, A, Smith, K, Chester, F, Knox, A, Stacey, A.R, Dukes, J, Baston, E, Griffin, S, Vuidepot, A, Jakobsen, B.K, Harper, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | TCRs with Distinct Specificity Profiles Use Different Binding Modes to Engage an Identical Peptide-HLA Complex.
J Immunol., 204, 2020
|
|
6EK9
 
 | | Cytosolic copper storage protein Csp from Streptomyces lividans: Cu loaded form | | Descriptor: | COPPER (I) ION, Cytosolic copper storage protein | | Authors: | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-09-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
5LYL
 
 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-2-[[(2~{S})-1-(1-adamantylamino)-3-cyclohexyl-1-oxidanylidene-propan-2-yl]sulfamoylamino]-6-azanyl-hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
6Y0K
 
 | |
