6YS5
 
 | | Acinetobacter baumannii ribosome-amikacin complex - 30S subunit head | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
6N6Q
 
 | |
5MVB
 
 | | Solution structure of a human G-Quadruplex hybrid-2 form in complex with a Gold-ligand. | | Descriptor: | DNA (26-MER), POTASSIUM ION, bis(m2-Oxo)-bis(2-methyl-2,2'-bipyridine)-di-gold(iii) | | Authors: | Wirmer-Bartoschek, J, Jonker, H.R.A, Bendel, L.E, Gruen, T, Bazzicalupi, C, Messori, L, Gratteri, P, Schwalbe, H. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Ligand/Hybrid-2-G-Quadruplex Complex Reveals Rearrangements that Affect Ligand Binding.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5J4C
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with cisplatin (soaked) and bound to mRNA and A-, P- and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Melnikov, S.V, Soll, D, Steitz, T.A, Polikanov, Y.S. | | Deposit date: | 2016-03-31 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into RNA binding by the anticancer drug cisplatin from the crystal structure of cisplatin-modified ribosome.
Nucleic Acids Res., 44, 2016
|
|
7QYN
 
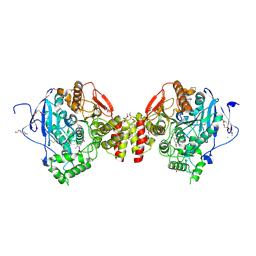 | | Mus musculus acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
6YNZ
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo composite tetramer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
6N2P
 
 | | Helical assembly of the CARD9 CARD | | Descriptor: | Caspase recruitment domain-containing protein 9 | | Authors: | Holliday, M.J, Rohou, A, Arthur, C.P, Dueber, E.C, Fairbrother, W.J. | | Deposit date: | 2018-11-13 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of autoinhibited and polymerized forms of CARD9 reveal mechanisms of CARD9 and CARD11 activation.
Nat Commun, 10, 2019
|
|
5MYY
 
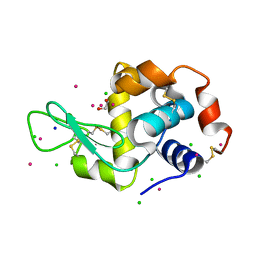 | | Hen Egg-White Lysozyme (HEWL) cocrystallized in the presence of Cadmium sulphate | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Panneerselvam, S, Burkhardt, A, Meents, A. | | Deposit date: | 2017-01-30 | | Release date: | 2017-07-12 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rapid cadmium SAD phasing at the standard wavelength (1 angstrom ).
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5MZ8
 
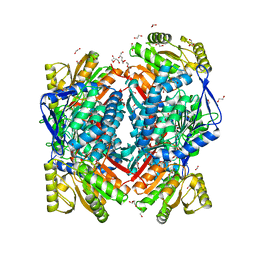 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with the reaction product succinate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-01-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
5N1J
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Giastas, P, Andreou, A, Balomenou, S, Bouriotis, V, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
7QGY
 
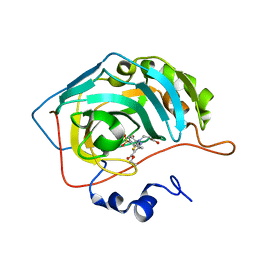 | | Human carbonic anhydrase II in complex with 3-((5-(ethoxycarbonyl)-4-methylthiazol-2-yl)(4-sulfamoylphenyl)amino)propanoic acid | | Descriptor: | 3-[(5-ethoxycarbonyl-4-methyl-1,3-thiazol-2-yl)-(4-sulfamoylphenyl)amino]propanoic acid, Carbonic anhydrase 2, ZINC ION | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Beta and Gamma Amino Acid-Substituted Benzenesulfonamides as Inhibitors of Human Carbonic Anhydrases.
Pharmaceuticals, 15, 2022
|
|
5N09
 
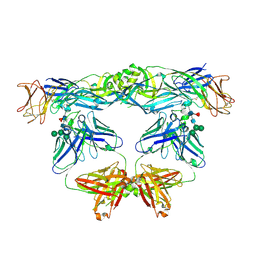 | | Crystal structure of L107C/A313C covalently linked dengue 2 virus envelope glycoprotein dimer in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 A11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 - Heavy chain, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 - Light chain, ... | | Authors: | Duquerroy, S, Rouvinski, A, Guardado-Calvo, P, Vaney, M.-C, Sharma, A, Rey, F. | | Deposit date: | 2017-02-02 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope.
Nat Commun, 8, 2017
|
|
7NYD
 
 | | cryoEM structure of 2C9-sMAC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Menny, A, Couves, E.C, Bubeck, D. | | Deposit date: | 2021-03-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis of soluble membrane attack complex packaging for clearance.
Nat Commun, 12, 2021
|
|
6YHR
 
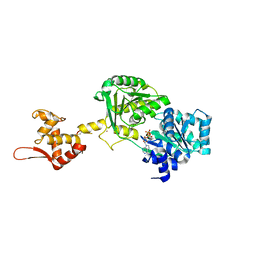 | | Crystal structure of Werner syndrome helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Werner syndrome ATP-dependent helicase, ZINC ION | | Authors: | Newman, J.A, Gavard, A.E, Savitsky, P, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the helicase core of Werner helicase, a key target in microsatellite instability cancers.
Life Sci Alliance, 4, 2021
|
|
6PLM
 
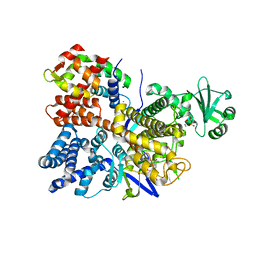 | | Legionella pneumophila SidJ/ Calmodulin 2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, Calmodulin-2, ... | | Authors: | Mao, Y, Sulpizio, A, Minelli, M.E, Wu, X. | | Deposit date: | 2019-07-01 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ.
Elife, 8, 2019
|
|
6VC2
 
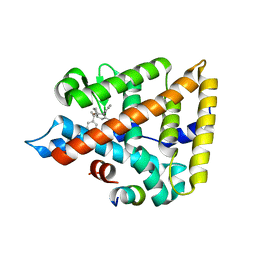 | | LRH-1 bound to SS-RJW100 and a fragment of the Tif2 Coactivator | | Descriptor: | (1S,3aS,6aS)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-ol, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Enantiomer-specific activities of an LRH-1 and SF-1 dual agonist.
Sci Rep, 10, 2020
|
|
5MZ4
 
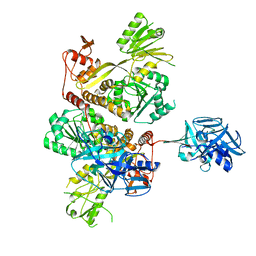 | | Crystal Structure of full-lengh CSFV NS3/4A | | Descriptor: | Genome polyprotein,Genome polyprotein | | Authors: | Tortorici, M.A, Rey, F.A. | | Deposit date: | 2017-01-30 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.048 Å) | | Cite: | A positive-strand RNA virus uses alternative protein-protein interactions within a viral protease/cofactor complex to switch between RNA replication and virion morphogenesis.
PLoS Pathog., 13, 2017
|
|
7T4G
 
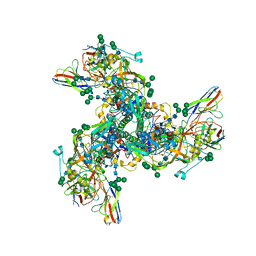 | |
8TBW
 
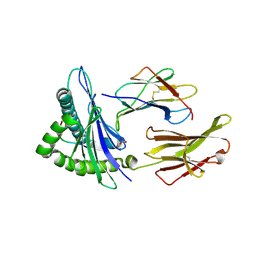 | | Human Class I MHC HLA-A2 in complex with sorting nexin 24 (127-135) peptide KLSHQPVLL | | Descriptor: | Beta-2-microglobulin, HLA-A*02:01 alpha chain, NITRATE ION, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
8U9G
 
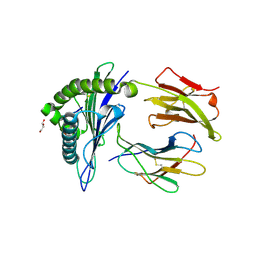 | | Human Class I MHC HLA-A2 bound to sorting nexin 24 (127-135) neoantigen KLSHQLVLL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-09-19 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
8TBV
 
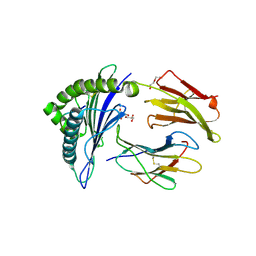 | | Human Class I MHC HLA-A2 in complex with sorting nexin 24 (127-135) neoantigen KLSHQLVLL | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
8U0Q
 
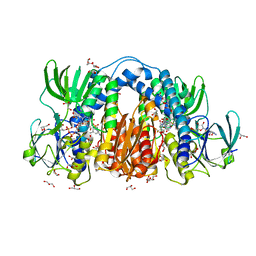 | | Co-crystal structure of optimized analog TDI-13537 provided new insights into the potency determinants of the sulfonamide inhibitor series | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dementiev, A.A, Michino, M, Vendome, J, Ginn, J, Bryk, R, Olland, A. | | Deposit date: | 2023-08-29 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Shape-Based Virtual Screening of a Billion-Compound Library Identifies Mycobacterial Lipoamide Dehydrogenase Inhibitors.
Acs Bio Med Chem Au, 3, 2023
|
|
5J55
 
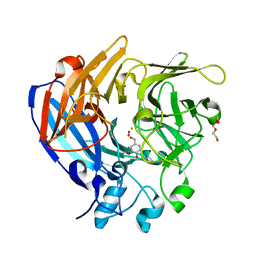 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Carotenoid oxygenase, FE (III) ION, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5NHG
 
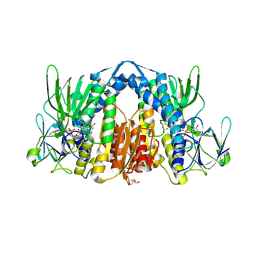 | | Crystal structure of the human dihydrolipoamide dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, ... | | Authors: | Szabo, E, Mizsei, R, Wilk, P, Zambo, Z, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structures of the disease-causing D444V mutant and the relevant wild type human dihydrolipoamide dehydrogenase.
Free Radic. Biol. Med., 124, 2018
|
|
6SVS
 
 | | Crystal Structure of U:A-U-rich RNA triple helix with 11 consecutive base triples | | Descriptor: | ADENOSINE-5'-PHOSPHATE-2',3'-CYCLIC PHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Ruszkowska, A, Ruszkowski, M, Hulewicz, J.P, Dauter, Z, Brown, J.A. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular structure of a U•A-U-rich RNA triple helix with 11 consecutive base triples.
Nucleic Acids Res., 48, 2020
|
|
