8OIO
 
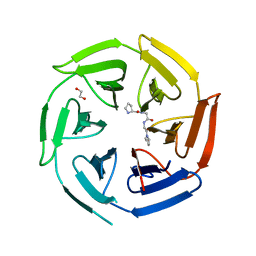 | | Crystal structure of the kelch domain of human KLHL12 in complex with PLEKHA4 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Kelch-like protein 12, ... | | Authors: | Dalietou, E.V, Chen, Z, Ramdass, A.E, Manning, C, Richardson, W, Aitmakhanova, K, Platt, M, Pike, A.C.W, Fedorov, O, Brennan, P, Bullock, A.N. | | Deposit date: | 2023-03-23 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Crystal structure of the kelch domain of human KLHL12 in complex with PLEKHA4 peptide
To Be Published
|
|
4WXL
 
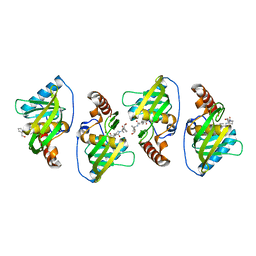 | |
5NVX
 
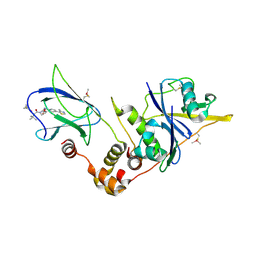 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-fluorocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 10) | | Descriptor: | Elongin-B, Elongin-C, N-[(1-fluorocyclopropyl)carbonyl]-3-methyl-L-valyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
6SCF
 
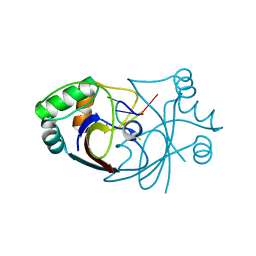 | | A viral anti-CRISPR subverts type III CRISPR immunity by rapid degradation of cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Athukoralage, J.S, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity.
Nature, 577, 2020
|
|
6SCV
 
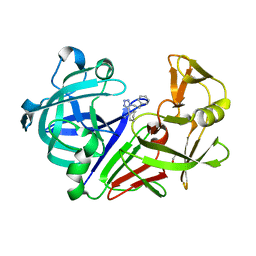 | | Endothiapepsin in complex with ligand 69 | | Descriptor: | Endothiapepsin, GLYCEROL, [(~{R})-cyclohexyl-[1-(2-phenylethyl)-1,2,3,4-tetrazol-5-yl]methyl]diazane | | Authors: | Magari, F, Heine, A, Klebe, G. | | Deposit date: | 2019-07-25 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Endothiapepsin in complex with ligand 69
To Be Published
|
|
5LWL
 
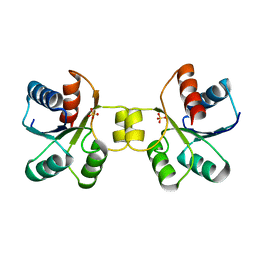 | | MaeR D54A mutant response regulator bound to sulfate | | Descriptor: | SULFATE ION, Transcriptional regulatory protein | | Authors: | Miguel-Romero, L, Casino, P, Landete, J.M, Monedero, V, Zuniga, M, Marina, A. | | Deposit date: | 2016-09-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The malate sensing two-component system MaeKR is a non-canonical class of sensory complex for C4-dicarboxylates.
Sci Rep, 7, 2017
|
|
7RCK
 
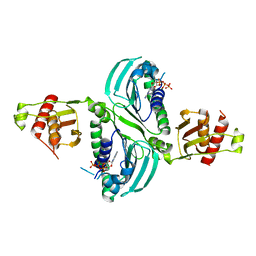 | | Crystal Structure of PMS2 with Substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
6XCO
 
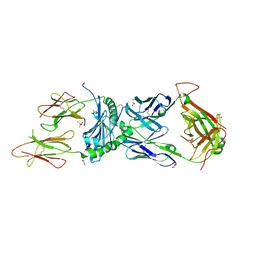 | | Immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, GLYCEROL, ... | | Authors: | Tran, T.M, Faridi, P, Lim, J.J, Ting, T.Y, Onwukwe, G, Bhattacharjee, P, Jones, M.C, Tresoldi, E, Cameron, J.F, La-Gruta, L.N, Purcell, W.A, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2020-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | T cell receptor recognition of hybrid insulin peptides bound to HLA-DQ8.
Nat Commun, 12, 2021
|
|
6BNX
 
 | | Crystal structure of V278E-glyoxalase I mutant from Zea mays in space group P6(3) | | Descriptor: | COBALT (II) ION, Lactoylglutathione lyase | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
5M26
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 in complex with methylhydroquinone | | Descriptor: | 2-methylbenzene-1,4-diol, FE (III) ION, Hydroquinone dioxygenase large subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
3ZKK
 
 | | Structure of the xylo-oligosaccharide specific solute binding protein from Bifidobacterium animalis subsp. lactis Bl-04 in complex with xylotetraose | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, XOS BINDING PROTEIN, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Ejby, M, Vujicic-Zagar, A, Fredslund, F, Svensson, B, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium Animalis Subsp. Lactis Bl-04
Mol.Microbiol., 90, 2013
|
|
6SXW
 
 | | Crystal structure of the first RRM domain of human Zinc finger protein 638 (ZNF638) | | Descriptor: | SULFATE ION, Zinc finger protein 638 | | Authors: | Newman, J.A, Aitkenhead, H, Wang, D, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-09-26 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Crystal structure of the first RRM domain of human Zinc finger protein 638 (ZNF638)
To Be Published
|
|
6V7M
 
 | |
5M33
 
 | | Structural tuning of CD81LEL (space group P21) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
4EB8
 
 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with DADMe-ImmG and phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, ... | | Authors: | Ho, M.C, Haapalainen, A.M, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-23 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
1W8N
 
 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, BACTERIAL SIALIDASE, SODIUM ION, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
8G8O
 
 | | The crystal structure of JAK2 in complex with Compound 31 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Tyrosine-protein kinase JAK2, ... | | Authors: | Miller, S.T, Ellis, D.A. | | Deposit date: | 2023-02-18 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Eyes on Topical Ocular Disposition: The Considered Design of a Lead Janus Kinase (JAK) Inhibitor That Utilizes a Unique Azetidin-3-Amino Bridging Scaffold to Attenuate Off-Target Kinase Activity, While Driving Potency and Aqueous Solubility.
J.Med.Chem., 66, 2023
|
|
7R70
 
 | | Crystal Structure of the UbArk2C fusion protein | | Descriptor: | GLYCEROL, Ubiquitin,E3 ubiquitin-protein ligase RNF165, ZINC ION | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
6LR6
 
 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
5MAY
 
 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PA14 IN COMPLEX WITH 2-Thiophenesulfonamide-N-(beta-L-fucopyranosyl methyl) | | Descriptor: | CALCIUM ION, Fucose-binding lectin PA-IIL, N-methyl-2-thiophenesulfonamide, ... | | Authors: | Sommer, R, Imberty, A, Titz, A, Varrot, A. | | Deposit date: | 2016-11-07 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Glycomimetic, Orally Bioavailable LecB Inhibitors Block Biofilm Formation of Pseudomonas aeruginosa.
J. Am. Chem. Soc., 140, 2018
|
|
5MBQ
 
 | | CeuE (H227A variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | Enterochelin uptake periplasmic binding protein | | Authors: | Wilde, E.J, Blagova, E.V, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
6PFZ
 
 | | Structure of a NAD-Dependent Persulfide Reductase from A. fulgidus | | Descriptor: | CALCIUM ION, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sazinsky, M.H, Shabdar, S, Garcia-Constineiras, A, Crane III, E.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.10003877 Å) | | Cite: | Structural and Kinetic Characterization of Hyperthermophilic NADH-Dependent Persulfide Reductase from Archaeoglobus fulgidus .
Archaea, 2021, 2021
|
|
6XQZ
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
3AQ3
 
 | |
6BYL
 
 | | Structure of 14-3-3 gamma bound to O-GlcNAcylated thr peptide | | Descriptor: | 14-3-3 protein gamma, 2-acetamido-2-deoxy-beta-D-glucopyranose, TSASTTVPVTTATTTTTSTW O-GlcNac peptide | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
