8SUJ
 
 | |
8SSJ
 
 | |
5FDP
 
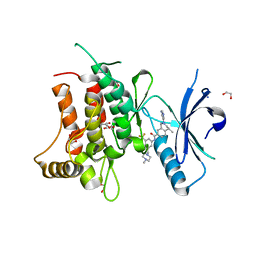 | | Structure of DDR1 receptor tyrosine kinase in complex with D2099 inhibitor at 2.25 Angstroms resolution. | | Descriptor: | (4~{S})-4-methyl-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinoline-7-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, Arrowsmith, C.H, von Delft, F, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Tetrahydroisoquinoline-7-carboxamides as Selective Discoidin Domain Receptor 1 (DDR1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
4U28
 
 | | Crystal structure of apo Phosphoribosyl isomerase A from Streptomyces sviceus ATCC 29083 | | Descriptor: | PHOSPHATE ION, Phosphoribosyl isomerase A | | Authors: | Chang, C, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
5OHX
 
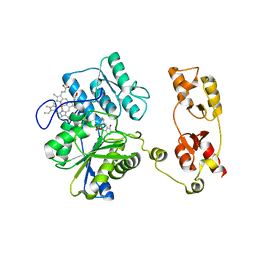 | | Structure of active cystathionine B-synthase from Apis mellifera | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gimenez-Mascarell, P, Majtan, T, Oyenarte, I, Ereno-Orbea, J, Majtan, J, Kraus, J.P, Klaudiny, J, Martinez-Cruz, L.A. | | Deposit date: | 2017-07-18 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of cystathionine beta-synthase from honeybee Apis mellifera.
J. Struct. Biol., 202, 2018
|
|
5FMF
 
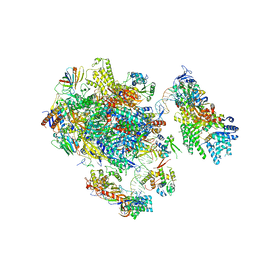 | | the P-lobe of RNA polymerase II pre-initiation complex | | Descriptor: | DNA REPAIR HELICASE RAD25, SSL2, DNA REPAIR HELICASE RAD3, ... | | Authors: | Murakami, K, Tsai, K, Kalisman, N, Bushnell, D.A, Asturias, F.J, Kornberg, R.D. | | Deposit date: | 2015-11-03 | | Release date: | 2015-11-25 | | Last modified: | 2017-07-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of an RNA Polymerase II Pre-Initiation Complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6TGN
 
 | |
5O89
 
 | | Crystal Structure of rsEGFP2 in the fluorescent on-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
4Z0Y
 
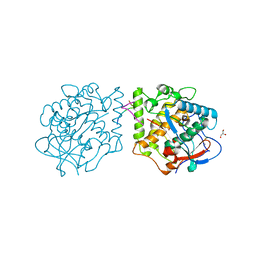 | | Active aurone synthase (polyphenol oxidase), copper B : sulfohistidine ~ 1.4 : 1 | | Descriptor: | Aurone synthase, COPPER (II) ION, GLYCEROL | | Authors: | Molitor, C, Mauracher, S.G, Rompel, A. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aurone synthase is a catechol oxidase with hydroxylase activity and provides insights into the mechanism of plant polyphenol oxidases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4U2K
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with anticancer compound at 2.13 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R)-2,3-diaminopropyl]-5-fluoropyrimidine-2,4(1H,3H)-dione, 1-[(2S)-2,3-diaminopropyl]-5-fluoropyrimidine-2,4(1H,3H)-dione, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with new anticancer compound at 1.17 A resolution
To Be Published
|
|
6FU5
 
 | | Structure of the kinase domain of human RIPK2 in complex with the inhibitor CSLP18 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, ~{N}-[5-[2-azanyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]-2-methoxy-phenyl]propane-1-sulfonamide | | Authors: | Pinkas, D.M, Bufton, J.C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Small molecule inhibitors reveal an indispensable scaffolding role of RIPK2 in NOD2 signaling.
EMBO J., 37, 2018
|
|
6YYZ
 
 | | tRNA-Guanine Transglycosylase (TGT) labeled with 5-fluorotryptophan in co-crystallized complex with 6-amino-2-(methylamino)-4-(2-((2R,3R,4R,5R)-3,4,5-trimethoxytetrahydrofuran-2-yl)ethyl)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-(methylamino)-4-(2-((2R,3R,4R,5R)-3,4,5-trimethoxytetrahydrofuran-2-yl)ethyl)-1H-imidazo[4,5-g]quinazolin-8(7H)-one, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Co-crystallization, 19F NMR and nanoESI-MS reveal dimer disturbing inhibitors and conformational changes at dimer contacts
To Be Published
|
|
4ZER
 
 | | Crystal structure of the Onc112 antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16s ribosomal RNA, 23s ribosomal RNA, ... | | Authors: | Seefeldt, A.C, Nguyen, F, Antunes, S, Perebaskine, N, Graf, M, Arenz, S, Inampudi, K.K, Douat, C, Guichard, G, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-04-20 | | Release date: | 2015-05-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6NXB
 
 | | ECAII IN COMPLEX WITH CITRATE AT PH 7 | | Descriptor: | CITRIC ACID, GLYCEROL, L-asparaginase 2 | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2019-02-08 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Opportunistic complexes of E. coli L-asparaginases with citrate anions.
Sci Rep, 9, 2019
|
|
5J25
 
 | | Endothiapepsin in complex with fragment 158 | | Descriptor: | 2-(4-methylpiperidin-1-yl)-N-[3-(propan-2-yl)-1,2-oxazol-5-yl]acetamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
5L8Q
 
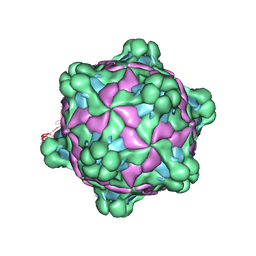 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-06-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
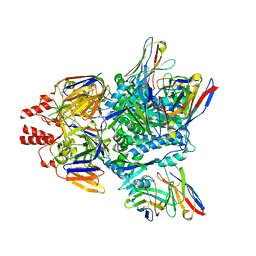
7LUE
 
 | |
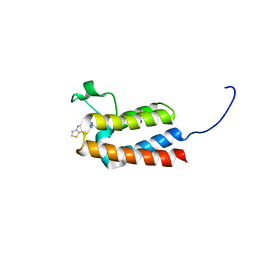
5L99
 
 | |
5FTG
 
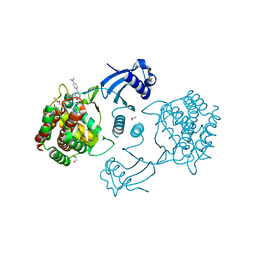 | | Human choline kinase a1 in complex with compound 1-[[4-[2-[4-[[4-(dimethylamino)pyridin-1- yl]methyl]phenoxy]ethoxy]phenyl]methyl]-N,N- dimethyl-pyridin-4-amine (compound 10a) | | Descriptor: | 1,2-ETHANEDIOL, 1-[[4-[2-[4-[[4-(dimethylamino)pyridin-1-yl]methyl]phenoxy]ethoxy]phenyl]methyl]-N,N-dimethyl-pyridin-4-amine, CHOLINE KINASE ALPHA | | Authors: | Schiaffino-Ortega, S, Baglioni, E, Mariotto, E, Bortolozzi, R, Serran-Aguilera, L, Rios-Marco, P, Carrasco-Jimenez, M.P, Gallo, M.A, Hurtado-Guerrero, R, Marco, C, Basso, G, Viola, G, Entrena, A, Lopez-Cara, L.C. | | Deposit date: | 2016-01-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design, Synthesis, Crystallization and Biological Evaluation of New Symmetrical Biscationic Compounds as Selective Inhibitors of Human Choline Kinase Alpha1 (Chokalpha1)
Sci.Rep., 6, 2016
|
|
6NX7
 
 | |
6NXE
 
 | |
5Z25
 
 | | Trimeric Alpha-Helix-Inserted Circular Permutant of Cytochrome c555 | | Descriptor: | Cytochrome c552, HEME C, TETRAETHYLENE GLYCOL | | Authors: | Oda, A, Nagao, S, Yamanaka, M, Ueda, I, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-12-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Construction of a Triangle-Shaped Trimer and a Tetrahedron Using an alpha-Helix-Inserted Circular Permutant of Cytochrome c555.
Chem Asian J, 13, 2018
|
|
7LUD
 
 | | Crystal structure of Fab ADI-14442 | | Descriptor: | ADI-14442 Fab Heavy chain, ADI-14442 Fab Light chain, SULFATE ION | | Authors: | Rush, S.A, McLellan, J.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein induces genetically and antigenically diverse antibody responses.
Immunity, 54, 2021
|
|
5Z37
 
 | |
5FLX
 
 | | Mammalian 40S HCV-IRES complex | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yamamoto, H, Collier, M, Loerke, J, Ismer, J, Schmidt, A, Hilal, T, Sprink, T, Yamamoto, K, Mielke, T, Burger, J, Shaikh, T.R, Dabrowski, M, Hildebrand, P.W, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Architecture of the Ribosome-Bound Hepatitis C Virus Internal Ribosomal Entry Site RNA.
Embo J., 34, 2015
|
|
