5TPZ
 
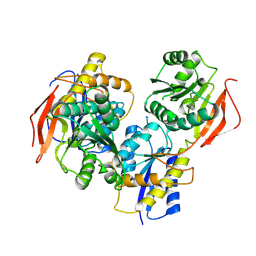 | | Crystal structure of amino terminal domains of the NMDA receptor subunit GluN1 and GluN2B in apo closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 2B, ... | | Authors: | Romero-Hernandez, A, Simorwski, N, Karakas, E, Furukawa, H. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.095 Å) | | Cite: | Molecular Basis for Subtype Specificity and High-Affinity Zinc Inhibition in the GluN1-GluN2A NMDA Receptor Amino-Terminal Domain.
Neuron, 92, 2016
|
|
3CZX
 
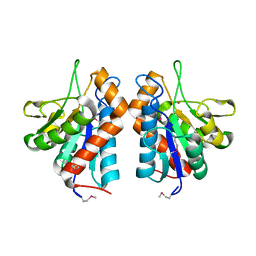 | |
5J8X
 
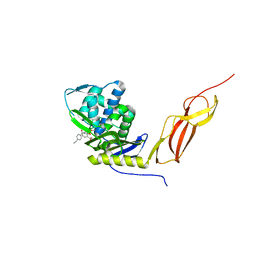 | | CRYSTAL STRUCTURE OF E. COLI PBP5 WITH 2C | | Descriptor: | (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, D-alanyl-D-alanine carboxypeptidase DacA | | Authors: | Brem, J, Cain, R, McDonough, M.A, Clifton, I.J, Fishwick, C.W.G, Schofield, C.J. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates.
Nat Commun, 7, 2016
|
|
4TT2
 
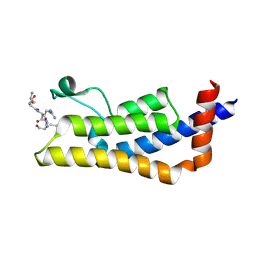 | | Crystal structure of ATAD2A bromodomain complexed with H4(1-20)K5Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4K5Ac | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
6GPA
 
 | | Beta-1,4-galactanase from Bacteroides thetaiotaomicron with galactose | | Descriptor: | Arabinogalactan endo-beta-1,4-galactanase, beta-D-galactopyranose | | Authors: | Hekelaar, J, Boger, M, Leeuwen van, S.S, Lammerts van Bueren, A, Dijkhuizen, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional characterization of a family GH53 beta-1,4-galactanase from Bacteroides thetaiotaomicron that facilitates degradation of prebiotic galactooligosaccharides.
J. Struct. Biol., 205, 2019
|
|
6M5N
 
 | | Apo-Form Structure of Borneol Dehydrogenase | | Descriptor: | ACETATE ION, Borneol dehydrogenase, SULFATE ION | | Authors: | Khine, A.A, Huang, K.F, Ko, T.P, Chen, H.P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural characterization of borneol dehydrogenase from Pseudomonas sp. TCU-HL1
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5TYI
 
 | | Grb7 SH2 with bicyclic peptide containing pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, Peptide inhibitor | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
2E27
 
 | | Crystal structure of Fv fragment of anti-ciguatoxin antibody complexed with ABC-ring of ciguatoxin | | Descriptor: | 3,7:6,10:9,14-TRIANHYDRO-2,5,11,12,13-PENTADEOXY-4-O-(METHOXYMETHYL)-L-ARABINO-L-ALLO-TETRADEC-12-ENONIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Yokota, A, Tanaka, Y, Tsumoto, K. | | Deposit date: | 2006-11-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Critical contribution of aromatic rings to specific recognition of polyether rings. The case of ciguatoxin CTX3C-ABC and its specific antibody 1C49.
J.Biol.Chem., 283, 2008
|
|
6H3G
 
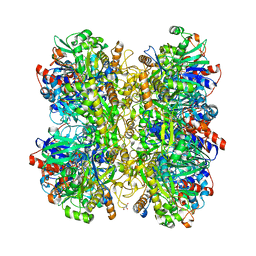 | | Alcohol oxidase from Phanerochaete chrysosporium | | Descriptor: | Alcohol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Nguyen, Q.-T, Romero, E, Dijkman, W.P, de Vasconcellos, S.P, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2018-07-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Engineering of Phanerochaete chrysosporium Alcohol Oxidase for Enhanced Oxidative Power toward Glycerol.
Biochemistry, 57, 2018
|
|
4H4K
 
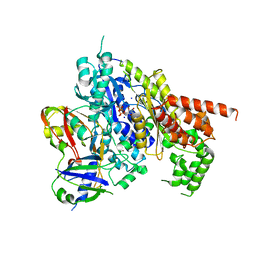 | | Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA-silencing complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cmr subunit Cmr2, CRISPR system Cmr subunit Cmr3, ... | | Authors: | Shao, Y, Cocozaki, A.I, Ramia, N.F, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2012-09-17 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structure of the cmr2-cmr3 subcomplex of the cmr RNA silencing complex.
Structure, 21, 2013
|
|
3M7B
 
 | |
8DQG
 
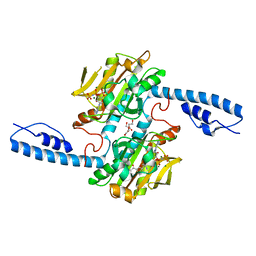 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to AMPPNP and acridone | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
6ZNV
 
 | | Protein polybromo-1 (PB1 BD2) Bound To DP28 | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperidin-4-ol, ACETATE ION, ... | | Authors: | Preuss, F, Mathea, S, Chatterjee, D, Wanior, M, Joerger, A.C, Knapp, S. | | Deposit date: | 2020-07-06 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
8E3U
 
 | |
6SFJ
 
 | | Crystal structure of p38 alpha in complex with compound 77 (MCP41) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-(2-methylphenyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
4LG8
 
 | | Crystal structure of PRPF19 WD40 repeats | | Descriptor: | Pre-mRNA-processing factor 19, SODIUM ION, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Tempel, W, He, H, Dobrovetsky, E, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the WD40 domain of human PRPF19.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
6F0S
 
 | |
5MTK
 
 | | Crystal structure of human Caspase-1 with (3S,6S,10aS)-N-((2S,3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-6-(isoquinoline-1-carboxamido)-5-oxodecahydropyrrolo[1,2-a]azocine-3-carboxamide (PGE-3935199) | | Descriptor: | (3~{S})-3-[[(3~{S},6~{S},10~{a}~{S})-6-(isoquinolin-1-ylcarbonylamino)-5-oxidanylidene-2,3,6,7,8,9,10,10~{a}-octahydro-1~{H}-pyrrolo[1,2-a]azocin-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2017-01-09 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Playing against the odds: scaffold hopping from 3D-fragments
To Be Published
|
|
8DQI
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to ATP and acridone after 2- weeks of crystal growth | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
5AF9
 
 | | Thrombin in complex with 4-Methoxy-N-(2-pyridinyl)benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methoxy-N-(pyridin-2-yl)benzamide, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-01-20 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
8AEL
 
 | | SYNJ2BP complex with a synthetic Vangl2 peptide (3mer). | | Descriptor: | CALCIUM ION, GLY-GLY-GLY-THR-SER-VAL, GLYCEROL, ... | | Authors: | Carrasco, K, Cousido Siah, A, Gogl, G, Betzi, S, McEwen, A, Kostmann, C, Trave, G. | | Deposit date: | 2022-07-13 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SYNJ2BP PDZ domain in complex with a synthetic Vangl2 peptide.
To Be Published
|
|
8Q7Y
 
 | |
5J31
 
 | | Crystal structure of 14-3-3zeta in complex with an alkyne cross-linked cyclic peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, Exoenzyme S | | Authors: | Wallraven, K, Cromm, P, Bier, D, Glas, A, Grossmann, T. | | Deposit date: | 2016-03-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Constraining an Irregular Peptide Secondary Structure through Ring-Closing Alkyne Metathesis.
Chembiochem, 17, 2016
|
|
8E3T
 
 | |
5MUW
 
 | | Atomic structure of P4 packaging enzyme fitted into a localized reconstruction of bacteriophage phi6 vertex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Packaging enzyme P4 | | Authors: | Sun, Z, El Omari, K, Sun, X, Ilca, S, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
