6YAA
 
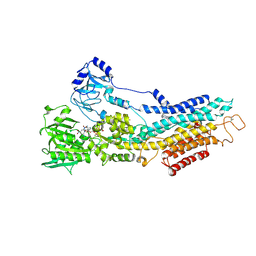 | | Structure of the (SR) Ca2+-ATPase bound to the inhibitor compound CAD204520 and TNP-ATP | | Descriptor: | 4-[2-[(2~{R})-2-[3-propyl-6-(trifluoromethyloxy)-1~{H}-indol-2-yl]piperidin-1-yl]ethyl]morpholine, POTASSIUM ION, SPIRO(2,4,6-TRINITROBENZENE[1,2A]-2O',3O'-METHYLENE-ADENINE-TRIPHOSPHATE, ... | | Authors: | Heit, S, Marchesini, M, Gherli, A, Montanaro, A, Patrizi, L, Sorrentino, C, Pagliaro, L, Rompietti, C, Kitara, S, Olesen, C.E, Moller, J.V, Savi, M, Bocchi, L, Vilella, R, Rizzi, F, Baglione, M, Rastelli, G, Loiacona, C, La Starza, R, Mecucci, C, Stegmair, K, Aversa, F, Stilli, D, Lund Winther, A.M, Sportoletti, P, Dalby-Brown, W, Roti, G, Bublitz, M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Blockade of Oncogenic NOTCH1 with the SERCA Inhibitor CAD204520 in T Cell Acute Lymphoblastic Leukemia.
Cell Chem Biol, 27, 2020
|
|
8GHB
 
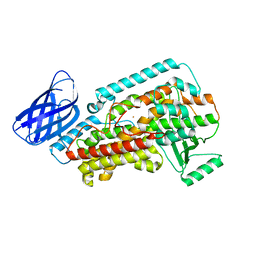 | | The structure of h12-LOX in monomeric form | | Descriptor: | FE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX12 | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
5Y9T
 
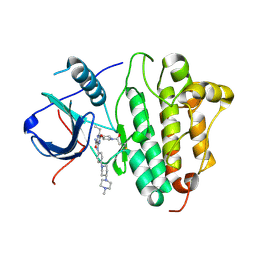 | | Crystal Structure of EGFR T790M mutant in complex with naquotinib | | Descriptor: | 6-ethyl-3-[[4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-5-[(3R)-1-prop-2-enoylpyrrolidin-3-yl]oxy-pyrazin e-2-carboxamide, Epidermal growth factor receptor | | Authors: | Mimasu, S, Tomimoto, Y, Maiko, I, Yasushi, A, Tatsuya, N. | | Deposit date: | 2017-08-28 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Pharmacological and Structural Characterizations of Naquotinib, a Novel Third-Generation EGFR Tyrosine Kinase Inhibitor, inEGFR-Mutated Non-Small Cell Lung Cancer.
Mol. Cancer Ther., 17, 2018
|
|
6M8O
 
 | |
5J5I
 
 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with 4-(2-amino-6-{bis[(pyridin-2-yl)methyl]amino}pyrimidin-4-yl)phenol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-amino-6-{bis[(pyridin-2-yl)methyl]amino}pyrimidin-4-yl)phenol, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Camacho Hernandez, G.A, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.326 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
6PNY
 
 | | X-ray Structure of Flpp3 | | Descriptor: | Flpp3 | | Authors: | Zook, J.D, Shekhar, M, Hansen, D.T, Conrad, C, Grant, T.D, Gupta, C, White, T, Barty, A, Basu, S, Zhao, Y, Zatsepin, N.A, Ishchenko, A, Batyuk, A, Gati, C, Li, C, Galli, L, Coe, J, Hunter, M, Liang, M, Weierstall, U, Nelson, G, James, D, Stauch, B, Craciunescu, F, Thifault, D, Liu, W, Cherezov, V, Singharoy, A, Fromme, P. | | Deposit date: | 2019-07-03 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | XFEL and NMR Structures of Francisella Lipoprotein Reveal Conformational Space of Drug Target against Tularemia.
Structure, 28, 2020
|
|
4RI4
 
 | |
4EMY
 
 | |
5J1O
 
 | | Excited state (Bound-like) sampled during RDC restrained Replica-averaged Metadynamics (RAM) simulations of the HIV-1 TAR complexed with cyclic peptide mimetic of Tat | | Descriptor: | Apical region (29mer) of the HIV-1 TAR element, Cyclic peptide mimetic of Tat | | Authors: | Borkar, A.N, Bardaro, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6LTM
 
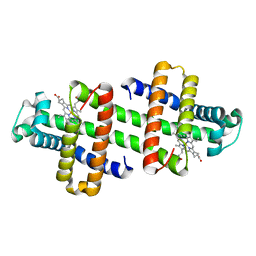 | | The dimeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
4YOA
 
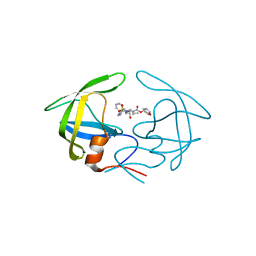 | | Crsystal structure HIV-1 Protease MDR769 L33F Complexed with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease | | Authors: | Kuiper, B.D, Keusch, B, Dewdney, T.G, Chordia, P, Brunzelle, J.S, Ross, K, Kovari, I.A, MacArthur, R, Salimnia, H, Kovari, L.C. | | Deposit date: | 2015-03-11 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The L33F darunavir resistance mutation acts as a molecular anchor reducing the flexibility of the HIV-1 protease 30s and 80s loops.
Biochem Biophys Rep, 2, 2015
|
|
1AE2
 
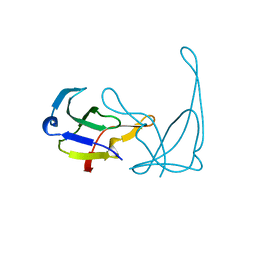 | | MUTANT L32R OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
6YC5
 
 | |
5ET8
 
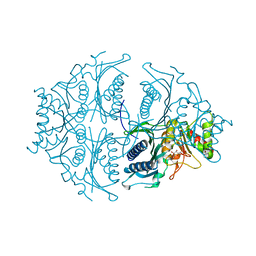 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | T-to-R switch of muscle FBPase involves extreme changes of secondary and quaternary structure
To Be Published
|
|
3FPK
 
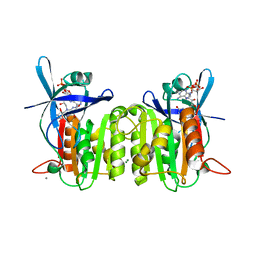 | | Crystal Structure of Ferredoxin-NADP Reductase from Salmonella typhimurium | | Descriptor: | CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase, ... | | Authors: | Kim, Y, Gu, M, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-05 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Ferredoxin-NADP Reductase from Salmonella typhimurium
To be Published
|
|
6AT4
 
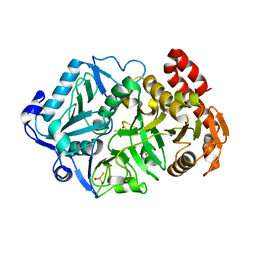 | |
4RWH
 
 | | Crystal structure of T cell costimulatory ligand B7-1 (CD80) | | Descriptor: | T-lymphocyte activation antigen CD80 | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | Deposit date: | 2014-12-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of T cell costimulatory ligand B7-1 (CD80)
To be Published
|
|
5EV3
 
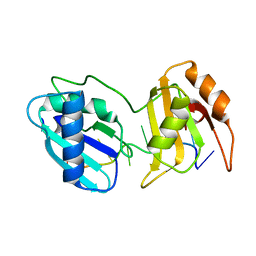 | |
7UZ1
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-bromo-valienide. | | Descriptor: | (1R,2S,3R,4R)-5-bromo-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Beta-galactosidase | | Authors: | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Karimi, R, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8DRD
 
 | | Ni(II)-bound B2 dimer (H60/H100/H104) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, HEME C, NICKEL (II) ION, ... | | Authors: | Choi, T.S, Tezcan, F.A. | | Deposit date: | 2022-07-20 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Design of a Flexible, Zn-Selective Protein Scaffold that Displays Anti-Irving-Williams Behavior.
J.Am.Chem.Soc., 144, 2022
|
|
2EHD
 
 | | Crystal Structure Analysis of Oxidoreductase | | Descriptor: | COBALT (II) ION, Oxidoreductase, short-chain dehydrogenase/reductase family | | Authors: | Kamiya, N, Hikima, T, Ebihara, A, Inoue, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-06 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of putative oxidoreductase from Thermus thermophilus HB8
to be published
|
|
6Y1Y
 
 | |
1XB4
 
 | |
7AUC
 
 | |
5MFI
 
 | | Designed armadillo repeat protein YIII(Dq.V2)4CqI in complex with peptide (KR)4 | | Descriptor: | (KR)4, YIII(Dq.V2)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
