6GOS
 
 | | E. coli Microcin synthetase McbBCD complex with pro-MccB17 bound | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin microcin B17, CHLORIDE ION, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-04 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
6F60
 
 | | The x-ray structure of bovine pancreatic ribonuclease in complex with a five-coordinate platinum(II) compound containing a sugar ligand | | Descriptor: | Ribonuclease pancreatic, SULFATE ION, five-coordinate platinum(II) compound | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2017-12-04 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Five-Coordinate Platinum(II) Compounds Containing Sugar Ligands: Synthesis, Characterization, Cytotoxic Activity, and Interaction with Biological Macromolecules.
Inorg Chem, 57, 2018
|
|
6F38
 
 | | Cryo-EM structure of two dynein tail domains bound to dynactin and HOOK3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Urnavicius, L, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
7JV2
 
 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody Fab fragment (local refinement of the receptor-binding motif and Fab variable domains) | | Descriptor: | S2H13 Fab heavy chain, S2H13 Fab light chain, Spike glycoprotein | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
6GQD
 
 | | Structure of human galactose-1-phosphate uridylyltransferase (GALT), with crystallization epitope mutations A21Y:A22T:T23P:R25L | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, Galactose-1-phosphate uridylyltransferase, ... | | Authors: | Fairhead, M, Strain-Damerell, C, Kopec, J, Bezerra, G.A, Zhang, M, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Structure of human galactose-1-phosphate uridylyltransferase (GALT), with crystallization epitope mutations A21Y:A22T:T23P:R25L
To Be Published
|
|
6F76
 
 | | Antibody derived (Abd-8) small molecule binding to KRAS. | | Descriptor: | 4-(2,3-dihydro-1,4-benzodioxin-5-yl)-~{N}-[3-[(dimethylamino)methyl]phenyl]-2-methoxy-aniline, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Bery, N, Cruz-Migoni, A, Quevedo, C.E, Phillips, S.V.E, Carr, S, Rabbitts, T.H. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions.
Elife, 7, 2018
|
|
6QJB
 
 | | Truncated Evasin-3 (tEv3 17-56) | | Descriptor: | Evasin-3 | | Authors: | Denisov, S.S, Ippel, J.H, Heinzman, A.C.A, Koenen, R.R, Ortega-Gomez, A, Soehnlein, O, Hackeng, T.M, Dijkgraaf, I. | | Deposit date: | 2019-01-24 | | Release date: | 2019-07-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Tick saliva protein Evasin-3 modulates chemotaxis by disrupting CXCL8 interactions with glycosaminoglycans and CXCR2.
J.Biol.Chem., 294, 2019
|
|
7RC0
 
 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-091-20 | | Descriptor: | 3C-like proteinase, 5-chloro-4-methylpyridin-3-yl 1H-indole-4-carboxylate, SODIUM ION | | Authors: | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
6QGP
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-0769 | | Descriptor: | 1-cycloheptyl-3-[3-(cyclopentyloxy)-4-methoxyphenyl]-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-0769
To be published
|
|
4YH2
 
 | |
6GYW
 
 | |
6FAP
 
 | | Crystal structure of human BAZ2A PHD zinc finger in complex with Fr23 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2017-12-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6GYY
 
 | |
6GIN
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with an Quinazolinone based ALK2 inhibitor with a 4-morpholinophenyl solvent accessible group. | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-morpholin-4-ylphenyl)-6-quinolin-4-yl-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Kopec, J, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
6QEC
 
 | | DNA binding domain of LUX ARRYTHMO in complex with DNA | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*GP*AP*AP*TP*AP*T*TP*AP*TP*AP*TP*TP*CP*GP*AP*A)-3'), GLYCEROL, Transcription factor LUX | | Authors: | Zubieta, C, Nayak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanisms of Evening Complex activity inArabidopsis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6GZ1
 
 | | Crystal Structure of the LeuO Effector Binding Domain | | Descriptor: | HTH-type transcriptional regulator LeuO, SULFATE ION | | Authors: | Fragel, S, Montada, A.M, Baumann, U, Schacherl, M, Schnetz, K. | | Deposit date: | 2018-07-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Characterization of the pleiotropic LysR-type transcription regulator LeuO of Escherichia coli.
Nucleic Acids Res., 47, 2019
|
|
6QEN
 
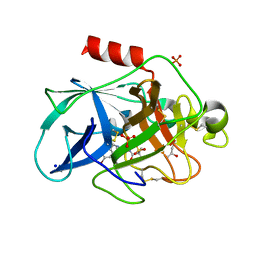 | | Crystal structure of Porcine Pancreatic Elastase (PPE) in complex with the 3-Oxo-beta-Sultam inhibitor LMC240 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-[[1-[(4-bromophenyl)methyl]-1,2,3-triazol-4-yl]methylamino]-2-methyl-1-oxidanylidene-propane-2-sulfonic acid, Chymotrypsin-like elastase family member 1, ... | | Authors: | Brito, J.A, Almeida, V.T, Carvalho, L.M, Moreira, R, Archer, M. | | Deposit date: | 2019-01-08 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.195 Å) | | Cite: | 3-Oxo-beta-sultam as a Sulfonylating Chemotype for Inhibition of Serine Hydrolases and Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
4TMK
 
 | |
5HCR
 
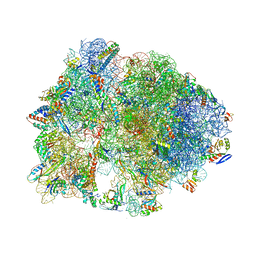 | | Crystal structure of antimicrobial peptide Oncocin 10wt bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
3J46
 
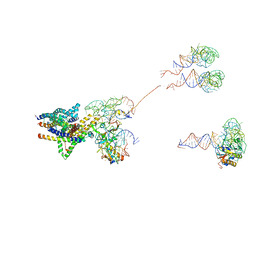 | | Structure of the SecY protein translocation channel in action | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L1, 50S ribosomal protein L23P, ... | | Authors: | Akey, C.W, Park, E, Menetret, J.F, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-23 | | Last modified: | 2019-07-03 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
8H6S
 
 | | Structure of acyltransferase VinK in complex with the loading acyl carrier protein of vicenistatin PKS | | Descriptor: | MAGNESIUM ION, Malonyl-CoA-[acyl-carrier-protein] transacylase, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Kawada, K, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Transient Interactions of Acyltransferase VinK with the Loading Acyl Carrier Protein of the Vicenistatin Modular Polyketide Synthase.
Biochemistry, 62, 2023
|
|
4TN0
 
 | | Crystal Structure of the C-terminal Periplasmic Domain of Phosphoethanolamine Transferase EptC from Campylobacter jejuni | | Descriptor: | UPF0141 protein yjdB, ZINC ION | | Authors: | Fage, C.D, Brown, D, Boll, J.M, Keatinge-Clay, A.T, Trent, M.S. | | Deposit date: | 2014-06-02 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic study of the phosphoethanolamine transferase EptC required for polymyxin resistance and motility in Campylobacter jejuni.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5H5K
 
 | | ATP and CMP bound Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Biswas, A, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2016-11-06 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of a hyperthermophilic thymidylate kinase enzyme reveal conformational substates along the reaction coordinate
FEBS J., 284, 2017
|
|
6V3U
 
 | | Crystal Structure of the NDM_FIM-1 like Metallo-beta-Lactamase from Erythrobacter litoralis in the Mono-Zinc Form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase II, ISOPROPYL ALCOHOL, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-26 | | Release date: | 2020-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the NDM_FIM-1 like Metallo-beta-Lactamase from Erythrobacter litoralis in the Mono-Zinc Form
To Be Published
|
|
6QFQ
 
 | | Structure of human Mcl-1 in complex with indole acid inhibitor | | Descriptor: | 7-(3,5-dimethyl-1~{H}-pyrazol-4-yl)-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
