2I0D
 
 | | Crystal structure of AD-81 complexed with wild type HIV-1 protease | | Descriptor: | (5S)-3-(3-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | Deposit date: | 2006-08-10 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
7ZQW
 
 | | Structure of the SARS-CoV-1 main protease in complex with AG7404 | | Descriptor: | 3C-like proteinase nsp5, ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | Authors: | Muriel-Goni, S, Fabrega-Ferrer, M, Herrera-Morande, A, Coll, M. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
3E4N
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 40 K: Laser off | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-08-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7T39
 
 | | Co-crystal structure of human PRMT9 in complex with MT221 inhibitor | | Descriptor: | 7-[5-S-(4-{[(2-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Protein arginine N-methyltransferase 9 | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Co-crystal structure of human PRMT9 in complex with MT221 inhibitor
To Be Published
|
|
6VU8
 
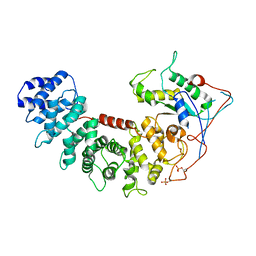 | | Structure of G-alpha-i bound to its chaperone Ric-8A | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | | Authors: | Seven, A.B, Hilger, D. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-18 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Cell Rep, 30, 2020
|
|
6PA5
 
 | |
4QUI
 
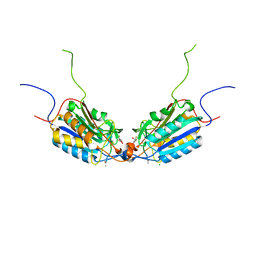 | | Caspase-3 F128AV266H | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CHLORIDE ION, ... | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
3EBG
 
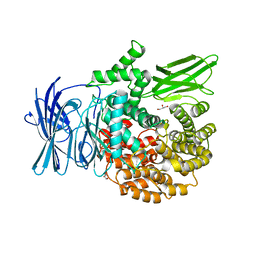 | | Structure of the M1 Alanylaminopeptidase from malaria | | Descriptor: | GLYCEROL, M1 family aminopeptidase, MAGNESIUM ION, ... | | Authors: | McGowan, S, Porter, C.J, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2008-08-27 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4QXX
 
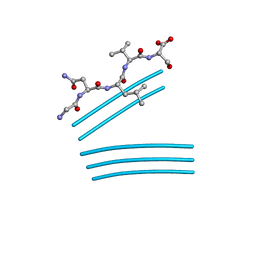 | |
5KHI
 
 | | HCN2 CNBD in complex with purine riboside-3', 5'-cyclic monophosphate (cPuMP) | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, Purine riboside-3',5'-cyclic monophosphate | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
7T88
 
 | | Crystal Structure of the C-terminal Domain of the Phosphate Acetyltransferase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IODIDE ION, ... | | Authors: | Kim, Y, Dementiev, A, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-15 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of c from Escherichia coli
To Be Published
|
|
1SSW
 
 | | Crystal structure of phage T4 lysozyme mutant Y24A/Y25A/T26A/I27A/C54T/C97A | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme | | Authors: | He, M.M, Baase, W.A, Xiao, H, Heinz, D.W, Matthews, B.W. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation
Protein Sci., 13, 2004
|
|
7SX3
 
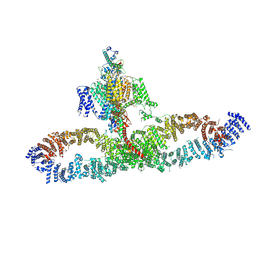 | | Human NALCN-FAM155A-UNC79-UNC80 channelosome with CaM bound, conformation 1/2 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Chua, H.C, Weidling, C, Chakouri, N, Noland, C.L, Schott, K, Chang, T, Tam, C, Patel, N, Arthur, C.P, Leitner, A, Ben-Johny, M, Ciferri, C, Pless, S.A, Payandeh, J. | | Deposit date: | 2021-11-22 | | Release date: | 2021-12-29 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural architecture of the human NALCN channelosome.
Nature, 603, 2022
|
|
2I81
 
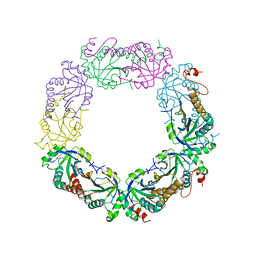 | | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced | | Descriptor: | 2-Cys Peroxiredoxin | | Authors: | Artz, J.D, Qiu, W, Dong, A, Lew, J, Ren, H, Zhao, Y, Kozieradski, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced
To be published
|
|
3EJ3
 
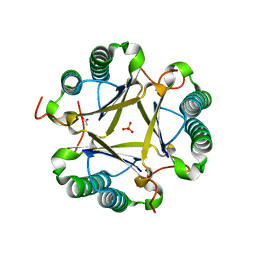 | | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity | | Descriptor: | ACETATE ION, Alpha-subunit of trans-3-chloroacrylic acid dehalogenase, Beta-subunit of trans-3-chloroacrylic acid dehalogenase, ... | | Authors: | Pegan, S, Serrano, H, Whitman, C.P, Mesecar, A.D. | | Deposit date: | 2008-09-17 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5CY1
 
 | |
2IAN
 
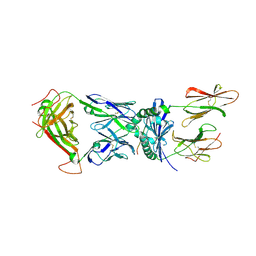 | | Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR | | Descriptor: | 15-mer peptide from Triosephosphate isomerase, CD4+ T cell receptor E8 alpha chain, CD4+ T cell receptor E8 beta chain, ... | | Authors: | Deng, L, Langley, R.J, Mariuzza, R.A. | | Deposit date: | 2006-09-08 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor
Nat.Immunol., 8, 2007
|
|
3ECZ
 
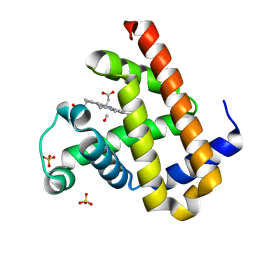 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser on [30 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-02 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6IEN
 
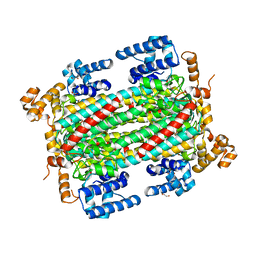 | | Substrate/product bound Argininosuccinate lyase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, ARGININOSUCCINATE, ... | | Authors: | Paul, A, Mishra, A, Surolia, A, Vijayan, M. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on M. tuberculosis argininosuccinate lyase and its liganded complex: Insights into catalytic mechanism.
IUBMB Life, 71, 2019
|
|
3EDB
 
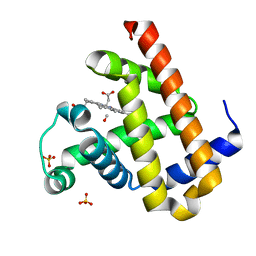 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser on [150 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-03 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1TLA
 
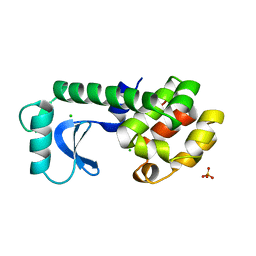 | | HYDROPHOBIC CORE REPACKING AND AROMATIC-AROMATIC INTERACTION IN THE THERMOSTABLE MUTANT OF T4 LYSOZYME SER 117 (RIGHT ARROW) PHE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, T4 LYSOZYME | | Authors: | Anderson, D.E, Hurley, J.H, Nicholson, H, Baase, W.A, Matthews, B.W. | | Deposit date: | 1993-03-22 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophobic core repacking and aromatic-aromatic interaction in the thermostable mutant of T4 lysozyme Ser 117-->Phe.
Protein Sci., 2, 1993
|
|
1T20
 
 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9-6I | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
5DIT
 
 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand (1R)-3-(3,4-dimethoxyphenyl)-1-f3-[2-(morpholin-4-yl)ethoxy]phenylgpropyl(2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate | | Descriptor: | (1R)-3-(3,4-dimethoxyphenyl)-1-{3-[2-(morpholin-4-yl)ethoxy]phenyl}propyl (2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Affinity Relationship Analysis of Selective FKBP51 Ligands.
J.Med.Chem., 58, 2015
|
|
1TLB
 
 | | Yeast coproporphyrinogen oxidase | | Descriptor: | Coproporphyrinogen III oxidase, SULFATE ION | | Authors: | Phillip, J.D, Whitby, F.G, Warby, C.A, Labbe, P, Yang, C, Pflugrath, J.W, Ferrara, J.D, Robinson, H, Kushner, J.P, Hill, C.P. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the oxygen-dependent coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
2WFA
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Beryllium trifluoride, in an open conformation. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
