8V7S
 
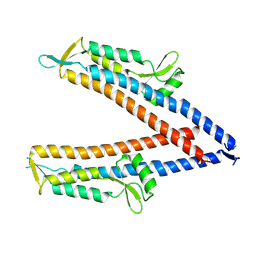 | | IpaD (122-321) Apo Structure | | Descriptor: | Invasin IpaD | | Authors: | Barker, S.A, Dickenson, N.E, Johnson, S.J, Morales, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural and functional characterization of the IpaD pi-helix reveals critical roles in DOC interaction, T3SS apparatus maturation, and Shigella virulence.
J.Biol.Chem., 300, 2024
|
|
5N4N
 
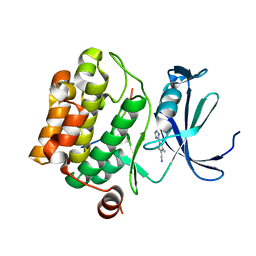 | | Crystal structure of human Pim-1 kinase in complex with a consensus peptide and fragment like molecule 3,4-dimethyl-5-(1H-1,2,4-triazol-3-yl)thiophene-2-carbonitrile | | Descriptor: | 3,4-dimethyl-5-(1~{H}-1,2,4-triazol-3-yl)thiophene-2-carbonitrile, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A crystallographic fragment study with human Pim-1 kinase
to be published
|
|
3UXY
 
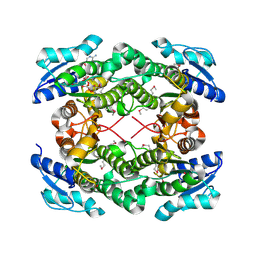 | | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase/reductase SDR | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides
To be Published
|
|
8KCL
 
 | |
8V4S
 
 | | Cryo-EM structure of the rat P2X7 receptor in the apo closed state purified in the absence of sodium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-DIPHOSPHATE, P2X purinoceptor 7, ... | | Authors: | Oken, A.C, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Glasfeld, A, Mansoor, S.E. | | Deposit date: | 2023-11-29 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | High-affinity agonism at the P2X 7 receptor is mediated by three residues outside the orthosteric pocket.
Nat Commun, 15, 2024
|
|
6QQC
 
 | |
1QX5
 
 | |
2WF5
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6-phosphate and trifluoromagnesate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Hounslow, A.M, Cliff, M.J, Williams, N.H, Hollfelder, F, Gamblin, S, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed by MgF-3 rather than by phosphoranes.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
8ENA
 
 | | Thaumatin native-SAD structure determined at 5 keV with a helium environmet | | Descriptor: | Thaumatin-1 | | Authors: | Karasawa, A, Andi, B, Ruchs, M.R, Shi, W, McSweeney, S, Hendrickson, W.A, Liu, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multi-crystal native-SAD phasing at 5 keV with a helium environment.
Iucrj, 9, 2022
|
|
6YPE
 
 | |
8EN9
 
 | | TehA native-SAD structure determined at 5 keV with a helium environment | | Descriptor: | CHLORIDE ION, SODIUM ION, Tellurite resistance protein TehA homolog, ... | | Authors: | Karasawa, A, Andi, B, Ruchs, M.R, Shi, W, McSweeney, S, Hendrickson, W.A, Liu, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Multi-crystal native-SAD phasing at 5 keV with a helium environment.
Iucrj, 9, 2022
|
|
7CMF
 
 | | Crystal structure of human P-cadherin REC12 (monomer) in complex with 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine (inhibitor) | | Descriptor: | 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3 | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulation of cadherin dimerization by chemical fragments as a trigger to inhibit cell adhesion
Commun Biol, 4, 2021
|
|
7RYX
 
 | | S. CEREVISIAE CYP51 COMPLEXED WITH VT-1129 | | Descriptor: | (2R)-2-(2,4-difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-{5-[4-(trifluoromethoxy)phenyl]pyridin-2-yl}propan-2-ol, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ruma, Y.N, Sagatova, A, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterisation of Candida parapsilosis CYP51 as a Drug Target Using Saccharomyces cerevisiae as Host.
J Fungi, 8, 2022
|
|
6MUX
 
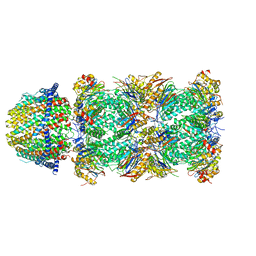 | | The structure of the Plasmodium falciparum 20S proteasome in complex with one PA28 activator | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Hanssen, E, Gillett, D.L, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2018-10-23 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol, 4, 2019
|
|
8GAD
 
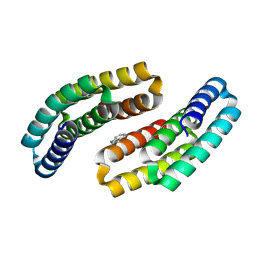 | |
8GAB
 
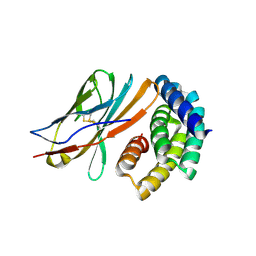 | | Crystal structure of CTLA-4 in complex with a high affinity CTLA-4 binder | | Descriptor: | CTLA-4 binder, Cytotoxic T-lymphocyte protein 4, POTASSIUM ION | | Authors: | Yang, W, Almo, S.C, Ghosh, A, Baker, D. | | Deposit date: | 2023-02-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
5VYM
 
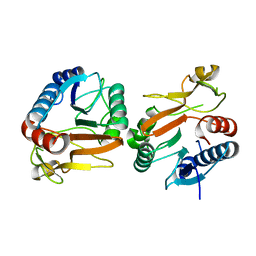 | | Crystal structure of beta-galactosidase from Bifidobacterium adolescentis | | Descriptor: | Beta-galactosidase BgaB | | Authors: | Chang, C, Cuff, M, Tesar, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-05-25 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Crystal structure of beta-galactosidase from Bifidobacterium adolescentis
To Be Published
|
|
7S65
 
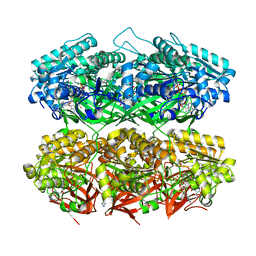 | | Compressed conformation of nighttime state KaiC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, ... | | Authors: | Sandate, C.R, Swan, J.A, Partch, C.L, Lander, G.C. | | Deposit date: | 2021-09-13 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Coupling of distant ATPase domains in the circadian clock protein KaiC.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S66
 
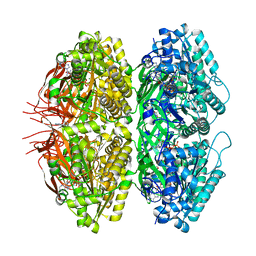 | | Extended conformation of nighttime state KaiC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Sandate, C.R, Swan, J.A, Partch, C.L, Lander, G.C. | | Deposit date: | 2021-09-13 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Coupling of distant ATPase domains in the circadian clock protein KaiC.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8EW4
 
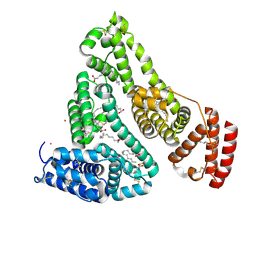 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 1 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Unciano, J, Lea, K, Kim, L, Lenkiewicz, J, Starban, I, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
5M1G
 
 | |
1KZV
 
 | |
6Y4V
 
 | | Crystal structure of p38 in complex with SR68 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(3-methylbutylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
1QYQ
 
 | | Crystal Structure of the cyclized S65G Y66G GFP variant | | Descriptor: | green-fluorescent protein | | Authors: | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-09-11 | | Release date: | 2003-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
8EY5
 
 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 3 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
