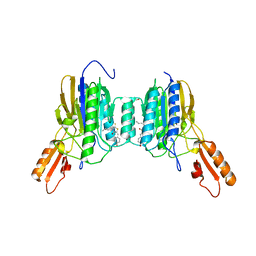
4Y8V
 
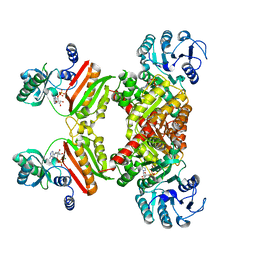 | |
6AS3
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | NHis AcrE1 protein | | Authors: | Shah, M, Calmettes, C, Pawluk, A, Mejdani, M, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
6ASC
 
 | | Mre11 dimer in complex with Endonuclease inhibitor PFM04 | | Descriptor: | (5E)-3-butyl-5-[(4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 1,2-ETHANEDIOL, MANGANESE (II) ION, ... | | Authors: | Moiani, D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Targeting Allostery with Avatars to Design Inhibitors Assessed by Cell Activity: Dissecting MRE11 Endo- and Exonuclease Activities.
Meth. Enzymol., 601, 2018
|
|
8OH5
 
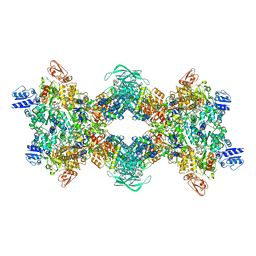 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 2) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
8OH9
 
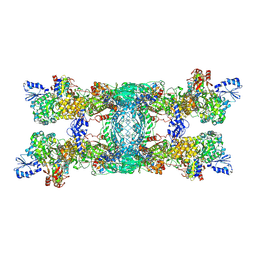 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 1) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Formate dehydrogenase-O, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
4Y6S
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, RC134, and manganese | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, MANGANESE (II) ION, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2015-02-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis and Bioactivity of beta-Substituted Fosmidomycin Analogues Targeting 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase.
J.Med.Chem., 58, 2015
|
|
7RNP
 
 | |
8AD6
 
 | | Structure of DarB bound to c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CBS domain-containing protein | | Authors: | Garcia-Pino, A, Talavera, A. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of DarB bound to c-di-AMP
To Be Published
|
|
8AHB
 
 | |
5U9T
 
 | | The Tris-thiolate Zn(II)S3Cl Binding Site Engineered by D-Cysteine Ligands in de Novo Three-stranded Coiled Coil Environment | | Descriptor: | ACETATE ION, CHLORIDE ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Ruckthong, L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-12-18 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | d-Cysteine Ligands Control Metal Geometries within De Novo Designed Three-Stranded Coiled Coils.
Chemistry, 23, 2017
|
|
7UIW
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4RG3
 
 | |
3FWO
 
 | | The large ribosomal subunit from Deinococcus radiodurans complexed with Methymycin | | Descriptor: | (3R,4S,5S,7R,9E,11S,12R)-12-ethyl-11-hydroxy-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 23S RIBOSOMAL RNA, 5S RIBOSOMAL RNA | | Authors: | Auerbach, T, Mermershtain, I, Bashan, A, Davidovich, C, Rozenberg, H, Sherman, D.H, Yonath, A. | | Deposit date: | 2009-01-19 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structural basis for the antibacterial activity of the 12-membered-ring mono-sugar macrolide methymycin
Biotechnologia, 1, 2009
|
|
6ESN
 
 | | Ligand complex of RORg LBD | | Descriptor: | (2~{R})-2-acetamido-~{N}-[4-(5-cyano-3-fluoranyl-2-methoxy-phenyl)thiophen-2-yl]-2-(4-ethylsulfonylphenyl)ethanamide, LYS-HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Nuclear receptor ROR-gamma, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2017-10-23 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Potent and Orally Bioavailable Inverse Agonists of ROR gamma t Resulting from Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6SBU
 
 | | X-ray Structure of Human LDHA with an Allosteric Inhibitor (Compound 3) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-[[4-[(5-chloranylthiophen-2-yl)carbonylamino]-1,3-bis(oxidanylidene)isoindol-2-yl]methyl]benzoic acid, L-lactate dehydrogenase A chain | | Authors: | Friberg, A, Puetter, V, Nguyen, D, Rehwinkel, H. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Evidence for Isoform-Selective Allosteric Inhibition of Lactate Dehydrogenase A.
Acs Omega, 5, 2020
|
|
6APQ
 
 | | Anti-Marburgvirus Nucleoprotein Single Domain Antibody B | | Descriptor: | Anti-Marburgvirus Nucleoprotein Single Domain Antibody B, CHLORIDE ION, SODIUM ION | | Authors: | Taylor, A.B, Garza, J.A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling a Drift Resistant Cryptotope withinMarburgvirusNucleoprotein Recognized by Llama Single-Domain Antibodies.
Front Immunol, 8, 2017
|
|
8SBG
 
 | |
8AHA
 
 | | rsEGFP2 photoswitched to its off-state at 100K | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Mantovanelli, A, Adam, V. | | Deposit date: | 2022-07-21 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Photophysical Studies at Cryogenic Temperature Reveal a Novel Photoswitching Mechanism of rsEGFP2.
J.Am.Chem.Soc., 145, 2023
|
|
5JED
 
 | | Apo-structure of humanised RadA-mutant humRadA28 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
7JXT
 
 | | Ovine COX-1 in complex with the subtype-selective derivative 2a | | Descriptor: | 2-[4,5-bis(2-chlorophenyl)-1H-imidazol-2-yl]-6-(prop-2-en-1-yl)phenyl methoxyacetate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ko, Y, Iaselli, M, Miciaccia, M, Friedrich, L, Schneider, G, Scilimati, A, Cingolani, G. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Learning from Nature: From a Marine Natural Product to Synthetic Cyclooxygenase-1 Inhibitors by Automated De Novo Design.
Adv Sci, 8, 2021
|
|
7UVE
 
 | | Drosophila melanogaster setdb1-tuor domain with peptide H3K9me2K14ac | | Descriptor: | Histone-lysine N-methyltransferase eggless, peptide H3K9me2K14ac | | Authors: | Zhou, M, Dong, A, Liu, K, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-01 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Drosophila melanogaster setdb1-tuor domain with peptide H3K9me2K14ac
To Be Published
|
|
7UJ0
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8DMY
 
 | | Cryo-EM structure of cardiac muscle alpha-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha cardiac muscle 1, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
6F7H
 
 | | Crystal structure of human AQP10 | | Descriptor: | Aquaporin-10, GLYCEROL, nonyl beta-D-glucopyranoside | | Authors: | Gotfryd, K, Wang, K, Missel, J.W, Pedersen, P.A, Gourdon, P. | | Deposit date: | 2017-12-08 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Human adipose glycerol flux is regulated by a pH gate in AQP10.
Nat Commun, 9, 2018
|
|
5X54
 
 | | Crystal structure of the Keap1 Kelch domain in complex with a tetrapeptide | | Descriptor: | ACE-GLU-TRP-TRP-TRP, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Sogabe, S, Kadotani, A, Lane, W, Snell, G. | | Deposit date: | 2017-02-14 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Kelch-like ECH-associated protein 1-inhibitory tetrapeptide and its structural characterization
Biochem. Biophys. Res. Commun., 486, 2017
|
|
