7PH7
 
 | | Nanodisc reconstituted MsbA in complex with nanobodies, spin-labeled at position T68C | | 分子名称: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,5-bis(oxidanyl)oxan-2-yl]oxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{R})-3-nonanoyloxytetradecanoyl]oxy-5-[[(3~{R})-3-octanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{S},5~{S},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanylnonanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-oxan-2-yl]methoxy]-5-oxidanyl-oxane-2-carboxylic acid, ATP-binding transport protein multicopy suppressor of htrB, ... | | 著者 | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | 登録日 | 2021-08-16 | | 公開日 | 2022-08-24 | | 最終更新日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
8CIE
 
 | | Crystal structure of the human CDKL5 kinase domain with compound YL-354 | | 分子名称: | 4-[[3,5-bis(fluoranyl)phenyl]carbonylamino]-~{N}-piperidin-4-yl-1~{H}-pyrazole-3-carboxamide, Cyclin-dependent kinase-like 5, SULFATE ION | | 著者 | Richardson, W, Chen, X, Newman, J.A, Bakshi, S, Lakshminarayana, B, Brooke, L, Bullock, A.N. | | 登録日 | 2023-02-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of a Potent and Selective CDKL5/GSK3 Chemical Probe That Is Neuroprotective.
Acs Chem Neurosci, 14, 2023
|
|
7B38
 
 | | Torpedo californica acetylcholinesterase complexed with Mg+2 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | 著者 | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L, Weiner, L. | | 登録日 | 2020-11-29 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
8ENL
 
 | | CryoEM structure of the high pH turnover-inactivated nitrogenase MoFe-protein | | 分子名称: | CHAPSO, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | 登録日 | 2022-09-30 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.37 Å) | | 主引用文献 | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
8ENN
 
 | | Homocitrate-deficient nitrogenase MoFe-protein from Azotobacter vinelandii nifV knockout | | 分子名称: | CHAPSO, CITRIC ACID, FE (III) ION, ... | | 著者 | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | 登録日 | 2022-09-30 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.58 Å) | | 主引用文献 | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
8ENM
 
 | | CryoEM structure of the high pH nitrogenase MoFe-protein under non-turnover conditions | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | 登録日 | 2022-09-30 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.14 Å) | | 主引用文献 | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
6P6K
 
 | |
7PPM
 
 | | SHP2 catalytic domain in complex with IRS1 (889-901) phosphopeptide (pSer-892, pTyr-896) | | 分子名称: | GLYCEROL, Insulin receptor substrate 1, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Sok, P, Zeke, A, Remenyi, A. | | 登録日 | 2021-09-14 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
8ENO
 
 | | Homocitrate-deficient nitrogenase MoFe-protein from A. vinelandii nifV knockout in complex with NafT | | 分子名称: | CHAPSO, CITRIC ACID, FE (III) ION, ... | | 著者 | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | 登録日 | 2022-09-30 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
8COY
 
 | | Structure of the catalytic domain of P. vivax Sub1 (triclinic crystal form) in complex with inhibitor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | 著者 | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | 登録日 | 2023-03-01 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.507 Å) | | 主引用文献 | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8CLR
 
 | | Integrated NMR/MD structure determination of a dynamic and thermodynamically stable CUUG RNA tetraloop | | 分子名称: | RNA hairpin with CUUG tetraloop | | 著者 | Oxenfarth, A, Kuemmerer, F, Bottaro, S, Schnieders, R, Pinter, G, Jonker, H.R.A, Fuertig, B, Richter, C, Blackledge, M, Lindorff-Larsen, K, Schwalbe, H. | | 登録日 | 2023-02-17 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Integrated NMR/Molecular Dynamics Determination of the Ensemble Conformation of a Thermodynamically Stable CUUG RNA Tetraloop.
J.Am.Chem.Soc., 145, 2023
|
|
7ARO
 
 | | Crystal structure of the non-ribose partial agonist LUF5833 bound to the adenosine A2A receptor | | 分子名称: | 2-azanyl-6-(1~{H}-imidazol-2-ylmethylsulfanyl)-4-phenyl-pyridine-3,5-dicarbonitrile, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | 著者 | Verdon, G, Amelia, T, van Veldhoven, J, Falsini, M, Liu, R, Heitman, L, van Westen, G, Segala, E, Cheng, R, Cooke, R, van der Es, D, Ijzerman, A. | | 登録日 | 2020-10-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.119 Å) | | 主引用文献 | Crystal Structure and Subsequent Ligand Design of a Nonriboside Partial Agonist Bound to the Adenosine A 2A Receptor.
J.Med.Chem., 64, 2021
|
|
8CP0
 
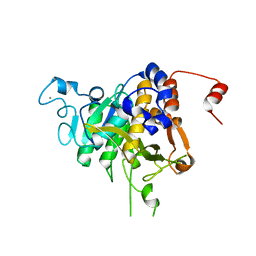 | | Structure of the catalytic domain of P. vivax Sub1 (trigonal crystal form) | | 分子名称: | CALCIUM ION, subtilisin | | 著者 | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | 登録日 | 2023-03-01 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.251 Å) | | 主引用文献 | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8COZ
 
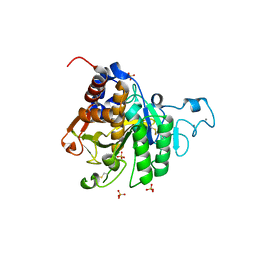 | | Structure of the catalytic domain of P. vivax Sub1 (triclinic crystal form) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | 著者 | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | 登録日 | 2023-03-01 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.438 Å) | | 主引用文献 | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1ABV
 
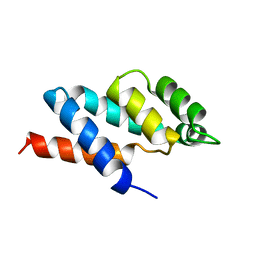 | | N-TERMINAL DOMAIN OF THE DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE FROM ESCHERICHIA COLI, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE | | 著者 | Wilkens, S, Dunn, S.D, Chandler, J, Dahlquist, F.W, Capaldi, R.A. | | 登録日 | 1997-01-29 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal domain of the delta subunit of the E. coli ATPsynthase.
Nat.Struct.Biol., 4, 1997
|
|
6PJI
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-43 | | 分子名称: | Protease NL4-3, SULFATE ION, methyl [(1S)-1-cyclopropyl-2-({(2S,3S,5S)-5-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-1,6-diphenylhexan-2-yl}amino)-2-oxoethyl]carbamate | | 著者 | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-06-28 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
8CMF
 
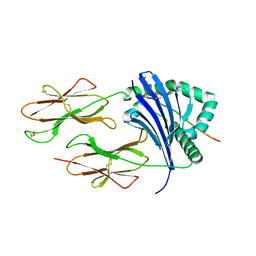 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 nsp3 epitope (orf1ab)1350-1364 | | 分子名称: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | 著者 | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | 登録日 | 2023-02-19 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
7PPL
 
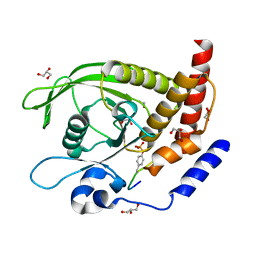 | | SHP2 catalytic domain in complex with IRS1 (625-639) phosphopeptide (pTyr-632, pSer-636) | | 分子名称: | ETHANOL, GLYCEROL, Insulin receptor substrate 1, ... | | 著者 | Sok, P, Zeke, A, Remenyi, A. | | 登録日 | 2021-09-14 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
7AOT
 
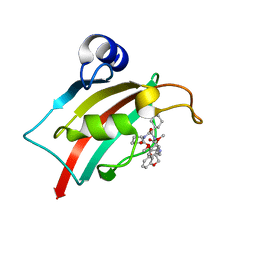 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone | | 分子名称: | (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | 著者 | Voll, A.M, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | 登録日 | 2020-10-15 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6MSY
 
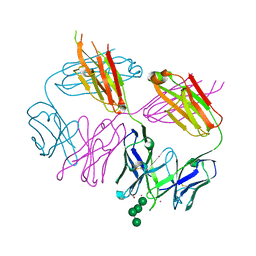 | | Anti-HIV-1 Fab Fab 2G12 + Man4 re-refinement | | 分子名称: | ACETATE ION, Fab 2G12, light chain, ... | | 著者 | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | 登録日 | 2018-10-18 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
6XJY
 
 | |
6XJQ
 
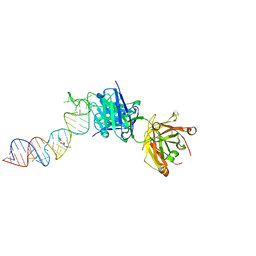 | | Crystal structure of a self-alkylating ribozyme - alkylated form with biotinylated epoxide substrate | | 分子名称: | 2-{[(4R)-4-hydroxyhexyl]oxy}ethyl 5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoate, Fab HAVx Heavy Chain, Fab HAVx Light Chain, ... | | 著者 | Koirala, D, Piccirilli, J.A. | | 登録日 | 2020-06-24 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.708 Å) | | 主引用文献 | Structural basis for substrate binding and catalysis by a self-alkylating ribozyme.
Nat.Chem.Biol., 18, 2022
|
|
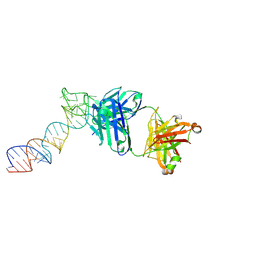
6XJZ
 
 | |
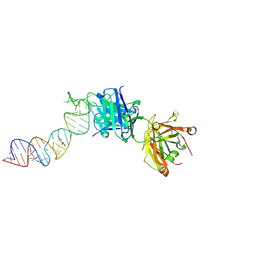
6XJW
 
 | |
7B62
 
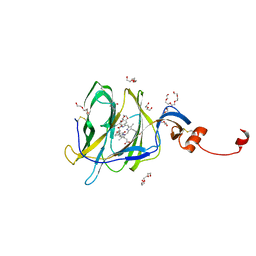 | | Crystal structure of SARS-CoV-2 spike protein N-terminal domain in complex with biliverdin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Pye, V.E, Rosa, A, Roustan, C, Cherepanov, P. | | 登録日 | 2020-12-07 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
