6WS6
 
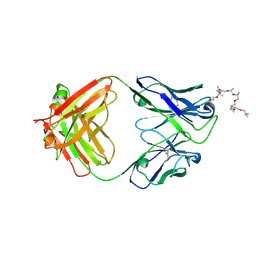 | | Structural and functional analysis of a potent sarbecovirus neutralizing antibody | | 分子名称: | O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), S309 antigen-binding (Fab) fragment, heavy chain, ... | | 著者 | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, Marco, A.D, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Telenti, A, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-04-30 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
4RSU
 
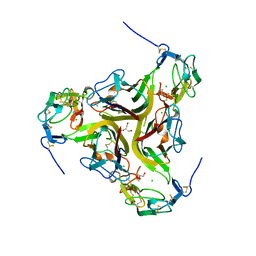 | | Crystal structure of the light and hvem complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | 著者 | Liu, W, Ramagoal, U.A, Himmel, D, Bonanno, J.B, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-11-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7OQ4
 
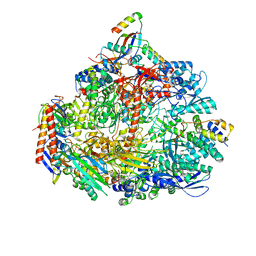 | | Cryo-EM structure of the ATV RNAP Inhibitory Protein (RIP) bound to the DNA-binding channel of the host's RNA polymerase | | 分子名称: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | 著者 | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | 登録日 | 2021-06-02 | | 公開日 | 2021-08-25 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
4S3F
 
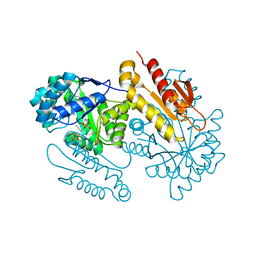 | | IspG in complex with Inhibitor 8 (compound 1077) | | 分子名称: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, FE3-S4 CLUSTER, but-3-yn-1-yl trihydrogen diphosphate | | 著者 | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | 登録日 | 2015-01-26 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4TMU
 
 | |
6WT4
 
 | | Structure of a bacterial STING receptor from Flavobacteriaceae sp. in complex with 3',3'-cGAMP | | 分子名称: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Bacterial STING, SULFATE ION | | 著者 | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | 登録日 | 2020-05-01 | | 公開日 | 2020-09-09 | | 最終更新日 | 2020-10-28 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
8AZW
 
 | | Cryo-EM structure of the plant 60S subunit | | 分子名称: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | 登録日 | 2022-09-06 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.14 Å) | | 主引用文献 | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
7P4M
 
 | |
7OMB
 
 | | Crystal structure of KOD DNA Polymerase in a ternary complex with a p/t duplex containing an extended 5' single stranded template overhang | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA polymerase, ... | | 著者 | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | 登録日 | 2021-05-21 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
7P53
 
 | |
6WU8
 
 | | Structure of human SHP2 in complex with inhibitor IACS-13909 | | 分子名称: | 1-[3-(2,3-dichlorophenyl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Leonard, P.G, Joseph, S, Rodenberger, A. | | 登録日 | 2020-05-04 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Allosteric SHP2 Inhibitor, IACS-13909, Overcomes EGFR-Dependent and EGFR-Independent Resistance Mechanisms toward Osimertinib.
Cancer Res., 80, 2020
|
|
7P1S
 
 | | Structure of KDNase from Trichophyton Rubrum in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | 分子名称: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, Extracellular sialidase/neuraminidase, SODIUM ION | | 著者 | Gloster, T.M, McMahon, S.A. | | 登録日 | 2021-07-02 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1F
 
 | | Structure of KDNase from Aspergillus terrerus in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | 分子名称: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, GLYCEROL, ... | | 著者 | Gloster, T.M, McMahon, S.A. | | 登録日 | 2021-07-01 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1O
 
 | |
4TTE
 
 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | 分子名称: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | 著者 | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | 登録日 | 2014-06-20 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
7P1U
 
 | |
7P1Q
 
 | |
7P76
 
 | | Re-engineered 2-deoxy-D-ribose-5-phosphate aldolase catalysing asymmetric Michael addition reactions, Schiff base complex with cinnamaldehyde | | 分子名称: | (2E)-3-phenylprop-2-enal, Deoxyribose-phosphate aldolase, GLYCEROL | | 著者 | Thunnissen, A.M.W.H, Rozeboom, H.J, Kunzendorf, A, Poelarends, G.J. | | 登録日 | 2021-07-19 | | 公開日 | 2021-10-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unlocking Asymmetric Michael Additions in an Archetypical Class I Aldolase by Directed Evolution.
Acs Catalysis, 11, 2021
|
|
7P1R
 
 | |
7P1D
 
 | |
7P1V
 
 | |
4RN5
 
 | | B1 domain of human Neuropilin-1 with acetate ion in a ligand-binding site | | 分子名称: | ACETATE ION, GLYCEROL, Neuropilin-1, ... | | 著者 | Allerston, C.K, Yelland, T.S, Jarvis, A, Jenkins, K, Winfield, N, Cheng, L, Jia, H, Zachary, I, Selwood, D.L, Djordjevic, S. | | 登録日 | 2014-10-23 | | 公開日 | 2015-10-28 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Conserved water molecules in a ligand-binding site of neuropilin-1
To be Published
|
|
7OQX
 
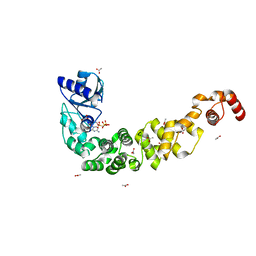 | | Crystal structure of a psychrophilic CCA-adding enzyme in complex with CMPcPP | | 分子名称: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ACETATE ION, CCA-adding enzyme, ... | | 著者 | Rollet, K, de Wijn, R, Bluhm, A, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | 登録日 | 2021-06-04 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus .
Comput Struct Biotechnol J, 19, 2021
|
|
6WEO
 
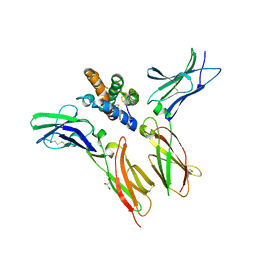 | | IL-22 Signaling Complex with IL-22R1 and IL-10Rbeta | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Saxton, R.A, Jude, K.M, Henneberg, L.T, Garcia, K.C. | | 登録日 | 2020-04-02 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The tissue protective functions of interleukin-22 can be decoupled from pro-inflammatory actions through structure-based design.
Immunity, 54, 2021
|
|
4RWO
 
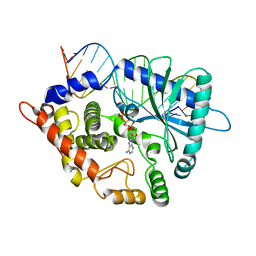 | | Crystal structure of the porcine OAS1 L149R mutant in complex with dsRNA and ApCpp in the AMP donor position | | 分子名称: | 2'-5'-oligoadenylate synthase 1, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | 著者 | Lohoefener, J, Steinke, N, Kay-Fedorov, P, Baruch, P, Nikulin, A, Tishchenko, S, Manstein, D.J, Fedorov, R. | | 登録日 | 2014-12-05 | | 公開日 | 2015-05-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Activation Mechanism of 2'-5'-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity.
Structure, 23, 2015
|
|
