6H75
 
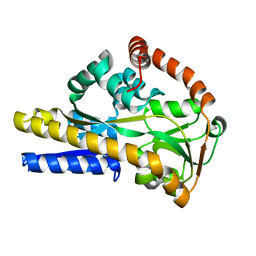 | | SiaP A11N in complex with Neu5Ac (RT) | | 分子名称: | N-acetyl-beta-neuraminic acid, Sialic acid-binding periplasmic protein SiaP | | 著者 | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | 登録日 | 2018-07-30 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
5KTH
 
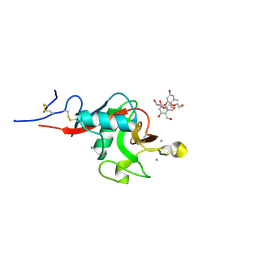 | | Structure of cow mincle complexed with brartemicin | | 分子名称: | 2,4-dihydroxy-6-methyl Benzoic acid, CALCIUM ION, TRIETHYLENE GLYCOL, ... | | 著者 | Feinberg, H, Rambaruth, N.D.S, Jegouzo, S.A.F, Jacobsen, K.M, Djurhuus, R, Poulsen, T.B, Taylor, M.E, Drickamer, K, Weis, W.I. | | 登録日 | 2016-07-11 | | 公開日 | 2016-08-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Binding Sites for Acylated Trehalose Analogs of Glycolipid Ligands on an Extended Carbohydrate Recognition Domain of the Macrophage Receptor Mincle.
J.Biol.Chem., 291, 2016
|
|
6F4U
 
 | |
6H7D
 
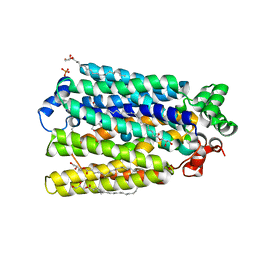 | | Crystal Structure of A. thaliana Sugar Transport Protein 10 in complex with glucose in the outward occluded state | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, PHOSPHATE ION, ... | | 著者 | Pedersen, B.P, Paulsen, P.A, Custodio, T.F. | | 登録日 | 2018-07-31 | | 公開日 | 2019-02-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the plant symporter STP10 illuminates sugar uptake mechanism in monosaccharide transporter superfamily.
Nat Commun, 10, 2019
|
|
5T0A
 
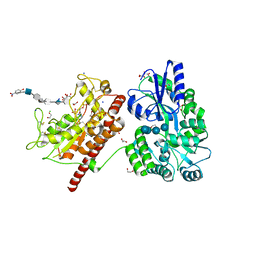 | | Crystal Structure of Heparan Sulfate 6-O-Sulfotransferase with bound PAP and heptasaccharide substrate | | 分子名称: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | 著者 | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | 登録日 | 2016-08-15 | | 公開日 | 2017-02-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
6B4E
 
 | | Crystal structure of Saccharomyces cerevisiae Gle1 CTD-Nup42 GBM complex | | 分子名称: | 1,2-ETHANEDIOL, Nucleoporin GLE1, Nucleoporin NUP42, ... | | 著者 | Lin, D.H, Correia, A.R, Cai, S.W, Huber, F.M, Jette, C.A, Hoelz, A. | | 登録日 | 2017-09-26 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Nat Commun, 9, 2018
|
|
6B4O
 
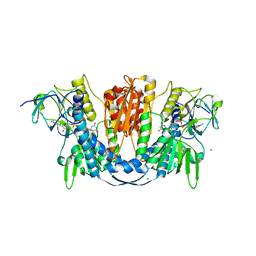 | | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, ... | | 著者 | Minasov, G, Warwzak, Z, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-09-27 | | 公開日 | 2017-10-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD.
To Be Published
|
|
7JKC
 
 | | Sheep Connexin-46 at 1.9 angstroms resolution by CryoEM | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | 著者 | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | 登録日 | 2020-07-28 | | 公開日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (1.9 Å) | | 主引用文献 | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
6F85
 
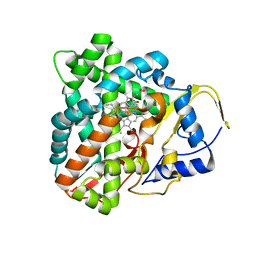 | |
4YKA
 
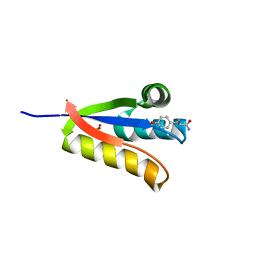 | | The structure of Agrobacterium tumefaciens ClpS2 in complex with L-tyrosinamide | | 分子名称: | ATP-dependent Clp protease adapter protein ClpS 2, L-TYROSINAMIDE, SULFATE ION | | 著者 | Stein, B, Grant, R.A, Sauer, R.T, Baker, T.A. | | 登録日 | 2015-03-04 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Structural Basis of an N-Degron Adaptor with More Stringent Specificity.
Structure, 24, 2016
|
|
6F8Y
 
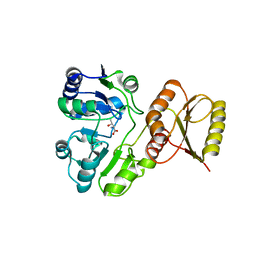 | | Crystal structure of P. abyssi Sua5 complexed with L-threonine | | 分子名称: | THREONINE, Threonylcarbamoyl-AMP synthase | | 著者 | Pichard-Kostuch, A, Zhang, W, Liger, D, Daugeron, M.C, Letoquart, J, Li de la Sierra-Gallay, I, Forterre, P, Collinet, B, van Tilbeurgh, H, Basta, T. | | 登録日 | 2017-12-13 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t6A tRNA modification.
RNA, 24, 2018
|
|
7JHE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with 2'-O-methylated m7GpppA Cap-1 and SAH Determined by Fixed-Target Serial Crystallography | | 分子名称: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | 著者 | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-20 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6B73
 
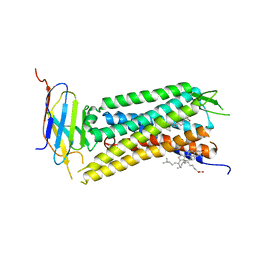 | | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor | | 分子名称: | CHOLESTEROL, N-[(5alpha,6beta)-17-(cyclopropylmethyl)-3-hydroxy-7,8-didehydro-4,5-epoxymorphinan-6-yl]-3-iodobenzamide, Nanobody, ... | | 著者 | Che, T, Majumdar, S, Zaidi, S.A, Kormos, C, McCorvy, J.D, Wang, S, Mosier, P.D, Uprety, R, Vardy, E, Krumm, B.E, Han, G.W, Lee, M.Y, Pardon, E, Steyaert, J, Huang, X.P, Strachan, R.T, Tribo, A.R, Pasternak, G.W, Carroll, I.F, Stevens, R.C, Cherezov, V, Katritch, V, Wacker, D, Roth, B.L. | | 登録日 | 2017-10-03 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor.
Cell, 172, 2018
|
|
6B74
 
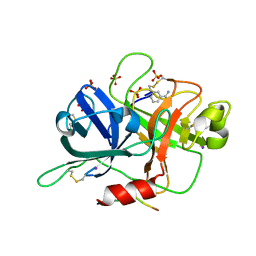 | |
8F1T
 
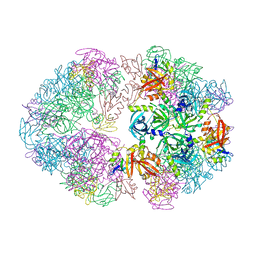 | | Structure of an 18mer DegP cage bound to the client protein hTRF1 | | 分子名称: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | 著者 | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | 登録日 | 2022-11-06 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (12.1 Å) | | 主引用文献 | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
4TLA
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Circadian clock protein kinase KaiC, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
6B7R
 
 | | Truncated strand 11-less green fluorescent protein | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Green fluorescent protein | | 著者 | Deng, A, Boxer, S.G. | | 登録日 | 2017-10-05 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Structural Insight into the Photochemistry of Split Green Fluorescent Proteins: A Unique Role for a His-Tag.
J. Am. Chem. Soc., 140, 2018
|
|
5L7P
 
 | | In silico-powered specific incorporation of photocaged Dopa at multiple protein sites | | 分子名称: | (2~{S})-2-azanyl-3-[3-[(2-nitrophenyl)methoxy]-4-oxidanyl-phenyl]propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Hauf, M, Richter, F, Schneider, T, Martins, B.M, Baumann, T, Durkin, P, Dobbek, H, Moeglich, A, Budisa, N. | | 登録日 | 2016-06-03 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Photoactivatable Mussel-Based Underwater Adhesive Proteins by an Expanded Genetic Code.
Chembiochem, 18, 2017
|
|
5HM9
 
 | | Crystal structure of MamO protease domain from Magnetospirillum magneticum (apo form) | | 分子名称: | MamO protease domain, poly(UNK) | | 著者 | Hershey, D.M, Ren, X, Hurley, J.H, Komeili, A. | | 登録日 | 2016-01-15 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | MamO Is a Repurposed Serine Protease that Promotes Magnetite Biomineralization through Direct Transition Metal Binding in Magnetotactic Bacteria.
Plos Biol., 14, 2016
|
|
7RPJ
 
 | | Cryo-EM structure of murine Dispatched NNN mutant | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | 著者 | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | 登録日 | 2021-08-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2021-11-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPH
 
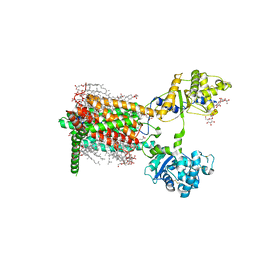 | | Cryo-EM structure of murine Dispatched 'R' conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | 著者 | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | 登録日 | 2021-08-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
6VBT
 
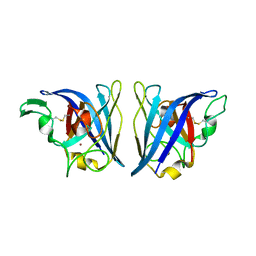 | |
6B8R
 
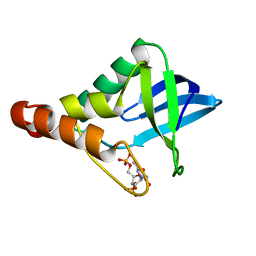 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23K/L36Q at cryogenic temperature | | 分子名称: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | 著者 | Robinson, A.C, Schlessman, J.L, Garcia-Moreno E, B, Benning, M. | | 登録日 | 2017-10-09 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Dielectric Properties of a Protein Probed by Reversal of a Buried Ion Pair.
J Phys Chem B, 122, 2018
|
|
4YFX
 
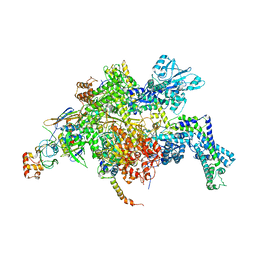 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | 登録日 | 2015-02-25 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.844 Å) | | 主引用文献 | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
2XZA
 
 | | Crystal Structure of recombinant A.17 antibody FAB fragment | | 分子名称: | FAB A.17 HEAVY CHAIN, FAB A.17 LIGHT CHAIN | | 著者 | Carletti, E, Nachon, F, Nicolet, Y, Masson, P, Kurkova, I, Smirnov, I, Friboulet, A, Tramontano, A, Gabibov, A. | | 登録日 | 2010-11-24 | | 公開日 | 2011-09-21 | | 最終更新日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Reactibodies Generated by Kinetic Selection Couple Chemical Reactivity with Favorable Protein Dynamics.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
