7M9Q
 
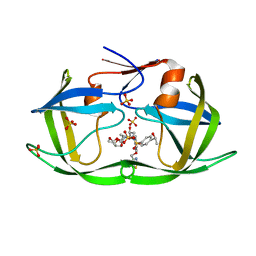 | | HIV-1 Protease WT (NL4-3) in Complex with LR4-33 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-[{4-[(1S)-1-hydroxyethyl]benzene-1-sulfonyl}(2-methylpropyl)amino]-1-{4-[(2-methyl-1,3-thiazol-4-yl)methoxy]phenyl}butan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | HIV-1 Protease WT (NL4-3) in Complex with LR4-33
To Be Published
|
|
5CV5
 
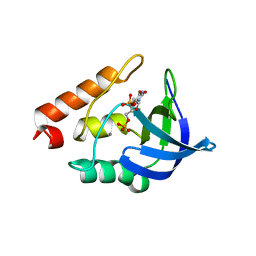 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS K64G/V66K/E67G at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Ortega Quintanilla, G, Robinson, A.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-07-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS K64G/V66K/E67G at cryogenic temperature
To be Published
|
|
7RWO
 
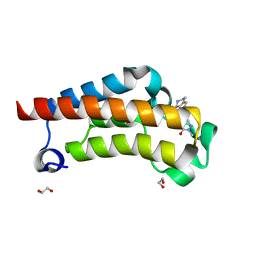 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
6GDI
 
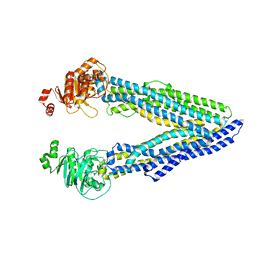 | | Structure of P-glycoprotein(ABCB1) in the post-hydrolytic state | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ford, R.C, Thonghin, N, Collins, R.F, Barbieri, A, Shafi, T, Siebert, A. | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Novel features in the structure of P-glycoprotein (ABCB1) in the post-hydrolytic state as determined at 7.9 angstrom resolution.
Bmc Struct.Biol., 18, 2018
|
|
7RPG
 
 | | X-ray crystal structure of OXA-24/40 K84D in complex with cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Powers, R.A, Mitchell, J.M, June, C.M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
7RPE
 
 | | X-ray crystal structure of OXA-24/40 in complex with ertapenem | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, BICARBONATE ION, Beta-lactamase, ... | | Authors: | Powers, R.A, Mitchell, J.M, June, C.M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
8F0M
 
 | | Monobody 12D5 bound to KRAS(G12D) | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Hattori, T, Glasser, E, Akkapeddi, P, Ketavarapu, G, Teng, K.W, Koide, A, Koide, S. | | Deposit date: | 2022-11-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Exploring switch II pocket conformation of KRAS(G12D) with mutant-selective monobody inhibitors.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1VAX
 
 | | Crystal Structure of Uricase from Arthrobacter globiformis | | Descriptor: | Uric acid oxidase | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2004-02-19 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Uricase from Arthrobacter globiformis
To be Published
|
|
7RPF
 
 | | X-ray crystal structure of OXA-24/40 in complex with doripenem | | Descriptor: | (2S,3R,4S)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, (4R,5S)-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-3-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-4,5-dihydro-1H-pyrrole-2-carboxylic acid, BICARBONATE ION, ... | | Authors: | Powers, R.A, Mitchell, J.M, June, C.M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
7TF4
 
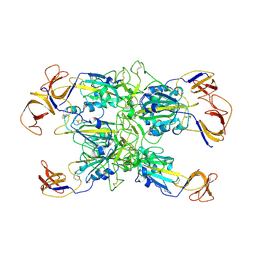 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) spike protein (focused refinement of RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
4PTT
 
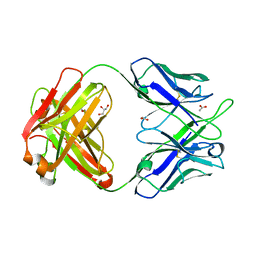 | | Crystal Structure of anti-23F strep Fab C05 | | Descriptor: | ACETATE ION, Antibody pn132p2C05, heavy chain, ... | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Smith, K, Schrader, J.W, Pai, E.F. | | Deposit date: | 2014-03-11 | | Release date: | 2015-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4N03
 
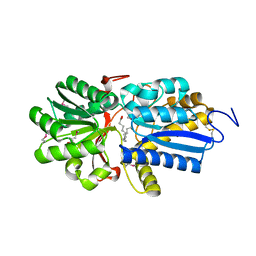 | | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata | | Descriptor: | 1,2-ETHANEDIOL, ABC-type branched-chain amino acid transport systems periplasmic component-like protein, PALMITIC ACID | | Authors: | Osipiuk, J, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata
To be Published
|
|
7RPD
 
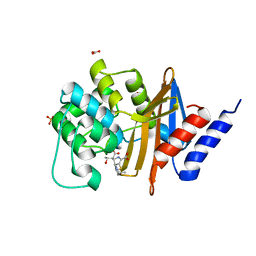 | | X-ray crystal structure of OXA-24/40 V130D in complex with ertapenem | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, BICARBONATE ION, Beta-lactamase, ... | | Authors: | Powers, R.A, Mitchell, J.M, June, C.M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
5N6N
 
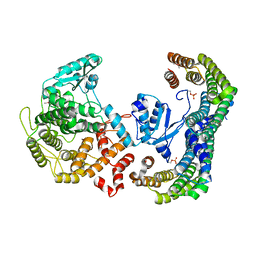 | | CRYSTAL STRUCTURE OF THE 14-3-3:NEUTRAL TREHALASE NTH1 COMPLEX | | Descriptor: | CALCIUM ION, Neutral trehalase, Protein BMH1, ... | | Authors: | Alblova, M, Smidova, A, Obsilova, V, Obsil, T. | | Deposit date: | 2017-02-15 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7M9K
 
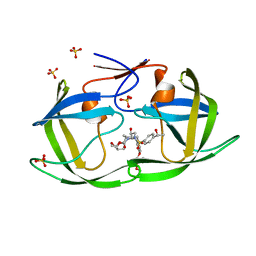 | | HIV-1 Protease WT (NL4-3) in Complex with LR3-48 | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-[{4-[(1R)-1-hydroxyethyl]benzene-1-sulfonyl}(2-methylpropyl)amino]butyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
5N2S
 
 | | Crystal structure of stabilized A1 receptor in complex with PSB36 at 3.3A resolution | | Descriptor: | 1-butyl-3-(3-oxidanylpropyl)-8-[(1~{R},5~{S})-3-tricyclo[3.3.1.0^{3,7}]nonanyl]-7~{H}-purine-2,6-dione, SULFATE ION, Soluble cytochrome b562,Adenosine receptor A1 | | Authors: | Cheng, R.K.Y, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-02-08 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
7TEX
 
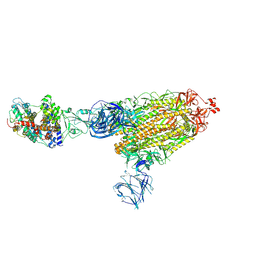 | | Cryo-EM structure of SARS-CoV-2 Delta (B.1.617.2) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
4IDT
 
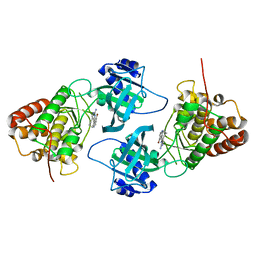 | | Crystal Structure of NIK with 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine (T28) | | Descriptor: | 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Liu, J, Sudom, A, Wang, Z. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibiting NF-KB-inducing kinase (NIK): Discovery, structure-based design, synthesis, structure activity relationship, and co-crystal structures
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7M9I
 
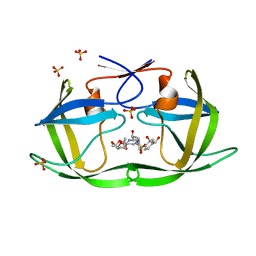 | | HIV-1 Protease (I84V) in Complex with LR2-26 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-ethylbutyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.817 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with LR2-26
To Be Published
|
|
7TE1
 
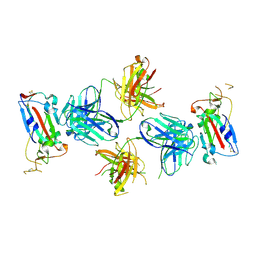 | |
5K2H
 
 | |
5N3U
 
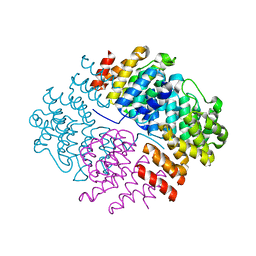 | | The structure of the complex of CpcE and CpcF of phycocyanin lyase from Nostoc sp. PCC7120 | | Descriptor: | Phycocyanobilin lyase subunit alpha, Phycocyanobilin lyase subunit beta | | Authors: | Hoeppner, A, Zhao, C, Xu, Q.-Z, Gaertner, W, Scheer, H, Zhao, K.-H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures and enzymatic mechanisms of phycobiliprotein lyases CpcE/F and PecE/F.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8P0L
 
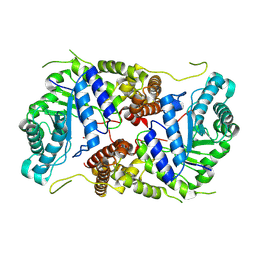 | | Crystal structure of human O-GlcNAcase in complex with an S-linked CKII peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, Protein O-GlcNAcase, ... | | Authors: | Males, A, Davies, G.J, Calvelo, M, Alteen, M.G, Vocadlo, D.J, Rovira, C. | | Deposit date: | 2023-05-10 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human O -GlcNAcase Uses a Preactivated Boat-skew Substrate Conformation for Catalysis. Evidence from X-ray Crystallography and QM/MM Metadynamics.
Acs Catalysis, 13, 2023
|
|
7MAB
 
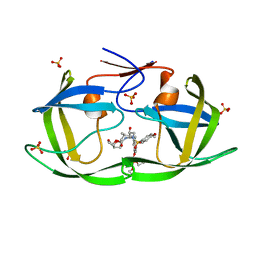 | | HIV-1 Protease (I84V) in Complex with GS-8374 | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with GS-8374
To Be Published
|
|
6VBS
 
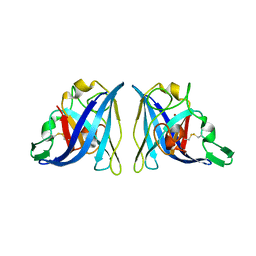 | |
