1U25
 
 | | Crystal structure of Selenomonas ruminantium phytase complexed with persulfated phytate in the C2221 crystal form | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
3NBP
 
 | | HIV-1 reverse transcriptase with aminopyrimidine inhibitor 2 | | Descriptor: | 4-(4-{[4-(4-cyano-2,6-dimethylphenoxy)pyrimidin-2-yl]amino}piperidin-1-yl)benzenesulfonamide, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Harris, S.F, Villasenor, A.G. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of piperidin-4-yl-aminopyrimidines as HIV-1 reverse transcriptase inhibitors. N-benzyl derivatives with broad potency against resistant mutant viruses.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6EO5
 
 | | Physcomitrella patens BBE-like 1 variant D396N | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PpBBE-like 1 D396N | | Authors: | Toplak, M, Winkler, A, Macheroux, P. | | Deposit date: | 2017-10-09 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The single berberine bridge enzyme homolog of Physcomitrella patens is a cellobiose oxidase.
FEBS J., 285, 2018
|
|
6Z2O
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, O-glycan protease, ZINC ION | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
4CPU
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[[4-[(3S)-3,4-dihydrothiophen-3-yl]phenyl]methyl]-2-[3-[(3Z,8S,11R)-11-oxidanyl-7,10-bis(oxidanylidene)-8-propan-2-yl-6,9-diazabicyclo[11.2.2]heptadeca-1(16),3,13(17),14-tetraen-11-yl]propyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
8Q5H
 
 | | Human KMN network (outer kinetochore) | | Descriptor: | Kinetochore protein Spc24, Kinetochore protein Spc25, Kinetochore scaffold 1, ... | | Authors: | Raisch, T, Polley, S, Vetter, I, Musacchio, A, Raunser, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the human KMN complex and implications for regulation of its assembly.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4CPS
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl [(2S)-1-(2-{3-[(3R,6S,10Z)-3-hydroxy-4,7-dioxo-6-(propan-2-yl)-5,8-diazabicyclo[11.2.2]heptadeca-1(15),10,13,16-tetraen-3-yl]propyl}-2-[4-(pyridin-3-yl)benzyl]hydrazinyl)-3,3-dimethyl-1-oxobutan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
6Z63
 
 | | FtsE structure from Streptococus pneumoniae in complex with ADP at 1.57 A resolution (spacegroup P 21) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE | | Authors: | Alcorlo, M, Straume, D, Havarstein, L.S, Hermoso, J.A. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
6ZLA
 
 | |
6ECH
 
 | | Pyruvate Kinase Isoform L-type with phosphorylated Ser12 (pS12) in complex with FBP | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Padyana, A, Tong, S. | | Deposit date: | 2018-08-07 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Distinct Hepatic PKA and CDK Signaling Pathways Control Activity-Independent Pyruvate Kinase Phosphorylation and Hepatic Glucose Production.
Cell Rep, 29, 2019
|
|
3AE7
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with 2-Iodo-N-(3-isopropoxy-phenyl)-benzamide | | Descriptor: | 2-iodo-N-[3-(1-methylethoxy)phenyl]benzamide, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
3A8H
 
 | | Crystal structure of Nitrile Hydratase mutant S113A complexed with Trimethylacetamide | | Descriptor: | 2,2-dimethylpropanamide, FE (III) ION, Nitrile hydratase subunit alpha, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
6ZD5
 
 | |
6EDH
 
 | | Taurine:2OG dioxygenase (TauD) bound to the vanadyl ion, taurine, and succinate | | Descriptor: | 2-AMINOETHANESULFONIC ACID, ACETATE ION, Alpha-ketoglutarate-dependent taurine dioxygenase, ... | | Authors: | Davis, K.M, Altmyer, M, Boal, A.K. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73000407 Å) | | Cite: | Structure of a Ferryl Mimic in the Archetypal Iron(II)- and 2-(Oxo)-glutarate-Dependent Dioxygenase, TauD.
Biochemistry, 58, 2019
|
|
2PEN
 
 | | Crystal structure of RbcX, crystal form I | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
4MES
 
 | |
1UGS
 
 | | Crystal structure of aY114T mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
4CWS
 
 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 2-{[2-amino-5-(1,3-benzodioxol-5-ylmethyl)[1,2,4]triazolo[1,5-c]quinazolin-8-yl]amino}ethanol, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
1KQN
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XENON | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human Nicotinamide/Nicotonic Acid Mononucleotide Adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2003
|
|
3NM9
 
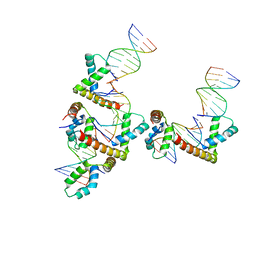 | | HMGD(M13A)-DNA complex | | Descriptor: | DNA 5'-D(*G*GP*CP*GP*AP*TP*AP*TP*CP*GP*C)-3', High mobility group protein D | | Authors: | Churchill, M.E.A, Klass, J, Zoetewey, D.L. | | Deposit date: | 2010-06-22 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural analysis of HMGD-DNA complexes reveals influence of intercalation on sequence selectivity and DNA bending.
J.Mol.Biol., 403, 2010
|
|
4LRK
 
 | |
3ACW
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with BPH-651 | | Descriptor: | (3R)-3-biphenyl-4-yl-1-azabicyclo[2.2.2]octan-3-ol, Dehydrosqualene synthase | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-13 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6EHG
 
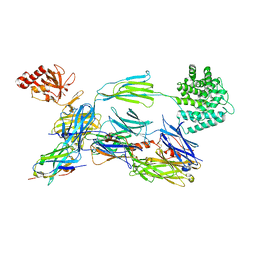 | | complement component C3b in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jensen, R.K, Andersen, K.R, Gadeberg, T.A.F, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-09-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A potent complement factor C3-specific nanobody inhibiting multiple functions in the alternative pathway of human and murine complement.
J. Biol. Chem., 293, 2018
|
|
4D53
 
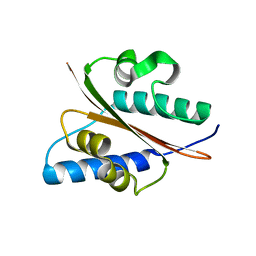 | | Outer surface protein BB0689 from Borrelia burgdorferi | | Descriptor: | BB0689 | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2014-11-02 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Analysis of Bb0689 from Borrelia Burgdorferi, a Member of the Bacterial CAP Superfamily.
J.Struct.Biol., 192, 2015
|
|
6EHQ
 
 | | E. coli Hydrogenase-2 (as isolated form). | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2017-09-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
