9CMJ
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease drug resistant mutant (L50F, E166V) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Coates, L, Gerlits, O. | | Deposit date: | 2024-07-15 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of SARS-CoV-2 Main Protease Mutations at Positions L50, E166, and L167 Rendering Resistance to Covalent and Noncovalent Inhibitors.
J.Med.Chem., 67, 2024
|
|
8EDQ
 
 | | E. coli pyruvate kinase (PykF) I264F | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pyruvate kinase, SULFATE ION | | Authors: | Donovan, K.A, Coombes, D, Dobson, R.C.J, Cooper, T.F. | | Deposit date: | 2022-09-05 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Beneficial mutations occurring in E. coli pyruvate kinase afford new allosteric mechanisms leading to faster resumption of growth
To Be Published
|
|
8QHO
 
 | | Human Carbonic Anhydrase II in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol | | Descriptor: | 1,1-bis(oxidanyl)-3,4-dihydro-2,1$l^{4}-benzoxaborinine, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-09-08 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Human Carbonic Anhydrase II in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol
To Be Published
|
|
9FZP
 
 | | Pseudomonas aeruginosa penicillin binding protein 3 in complex with cefepime | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-05 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
9EO4
 
 | | Outward-open structure of human dopamine transporter bound to cocaine | | Descriptor: | CHLORIDE ION, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nielsen, J.C, Salomon, K, Kalenderoglou, I.E, Bargmeyer, S, Pape, T, Shahsavar, A, Loland, C.J. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the human dopamine transporter in complex with cocaine.
Nature, 632, 2024
|
|
9GRQ
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-12 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9FVD
 
 | | Cryo-EM structure of single-layered nucleoprotein-RNA helical assembly from Marburg virus, trimeric repeat unit | | Descriptor: | Nucleoprotein, RNA (5'-R(P*AP*GP*AP*CP*AP*CP*AP*CP*AP*AP*AP*AP*AP*CP*AP*AP*GP*A)-3') | | Authors: | Zinzula, L, Beck, F, Camasta, M, Bohn, S, Liu, C, Morado, D, Bracher, A, Plitzko, J.M, Baumeister, W. | | Deposit date: | 2024-06-26 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of single-layered nucleoprotein-RNA complex from Marburg virus
Nat Commun, 2024
|
|
8QQ9
 
 | | human carbonic anhydrase I in complex with 1-benzyl-3-(1-hydroxy-3,4-dihydro-1H-benzo[c][1,2]oxaborinin-7-yl)thiourea | | Descriptor: | 1-[1,1-bis(oxidanyl)-3,4-dihydro-2,1$l^{4}-benzoxaborinin-7-yl]-3-(phenylmethyl)thiourea, Carbonic anhydrase 1, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | human carbonic anhydrase I in complex with 1-benzyl-3-(1-hydroxy-3,4-dihydro-1H-benzo[c][1,2]oxaborinin-7-yl)thiourea
To Be Published
|
|
8QQA
 
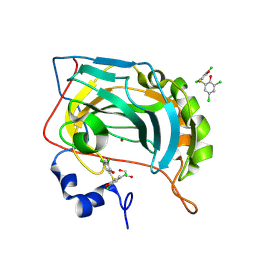 | |
8QTN
 
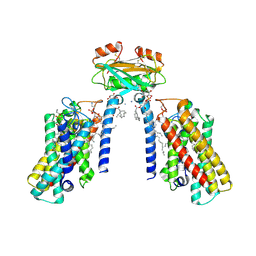 | | Cryo-EM structure of the apo yeast Ceramide Synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, AMMONIUM ION, Ceramide synthase LAC1, ... | | Authors: | Schaefer, J, Clausmeyer, L, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM Structure of the Yeast Ceramide Synthase Complex
To Be Published, 2024
|
|
8QUN
 
 | |
7L2Y
 
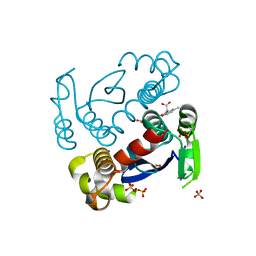 | |
7ODN
 
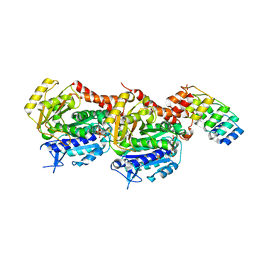 | | Crystal structure of TD1-mebendazole complex | | Descriptor: | CHLORIDE ION, Designed Ankyrin Repeat Protein (DARPIN) D1, GLYCEROL, ... | | Authors: | Oliva, M.A, Bonato, F, Diaz, J.F. | | Deposit date: | 2021-04-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Effect of Clinically Used Microtubule Targeting Drugs on Viral Infection and Transport Function.
Int J Mol Sci, 23, 2022
|
|
8QTR
 
 | | Cryo-EM structure of the FB-bound yeast Ceramide Synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, AMMONIUM ION, Ceramide synthase LAC1, ... | | Authors: | Schaefer, J, Clausmeyer, L, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-10-13 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structure of the Yeast Ceramide Synthase Complex
To Be Published, 2024
|
|
6OV3
 
 | |
6ZSH
 
 | |
4WDE
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation T311A | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, GLYCEROL | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WA7
 
 | | Crystal Structure of a GDP-bound Q61L Oncogenic Mutant of Human GT- Pase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Manandhar, A, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2014-08-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations.
Mol Cancer Res., 13, 2015
|
|
8EQ0
 
 | |
8EOM
 
 | | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973 | | Descriptor: | 4-(4-methylpiperazine-1-sulfonyl)benzamide, SULFATE ION, TP53-binding protein 1, ... | | Authors: | The, J, Hong, Z, Headey, S, Gunzburg, M, Doak, B, James, L.I, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-03 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973
to be published
|
|
8EQ3
 
 | | Escherichia coli pyruvate kinase A301T | | Descriptor: | IMIDAZOLE, MALONATE ION, Pyruvate kinase, ... | | Authors: | Donovan, K.A, Coombes, D, Dobson, R.C.J, Cooper, T.F. | | Deposit date: | 2022-10-07 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Beneficial mutations occurring in E. coli pyruvate kinase afford new allosteric mechanisms leading to faster resumption of growth
To Be Published
|
|
4WCI
 
 | | Crystal structure of the 1st SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide (aa 378-393) from human RIN3 | | Descriptor: | CD2-associated protein, Ras and Rab interactor 3, SULFATE ION | | Authors: | Rouka, E, Simister, P.C, Janning, M, Kirsch, K.H, Krojer, T, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-04 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
7KNV
 
 | | Solution NMR structure of CDHR3 extracellular domain EC1 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3 | | Authors: | Lee, W, Tonelli, M, Frederick, R.O, Watters, K.E, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2020-11-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Determination of the CDHR3 Rhinovirus-C Binding Domain, EC1
Viruses, 13, 2021
|
|
8EU4
 
 | |
6RFH
 
 | |
