4V0B
 
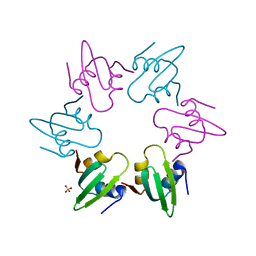 | | Escherichia coli FtsH hexameric N-domain | | Descriptor: | ATP-DEPENDENT ZINC METALLOPROTEASE FTSH, SULFATE ION | | Authors: | Serek-Heuberger, J, Martin, J, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2014-09-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and Evolution of N-Domains in Aaa Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
6LD1
 
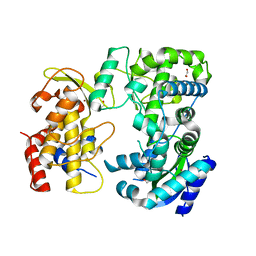 | | Zika NS5 polymerase domain | | Descriptor: | 1,2-ETHANEDIOL, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
6XF7
 
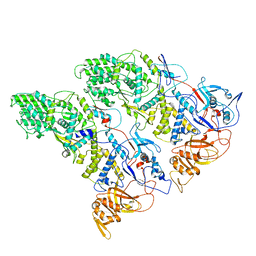 | | SLP | | Descriptor: | Lambda 1 protein | | Authors: | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
6LC7
 
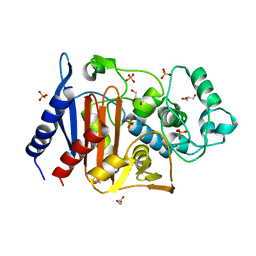 | | Crystal structure of AmpC Ent385 free form | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Beta-lactamase, GLYCEROL, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2019-11-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Reduced Susceptibility to Ceftazidime-Avibactam and Cefiderocol inEnterobacter cloacaeDue to AmpC R2 Loop Deletion.
Antimicrob.Agents Chemother., 64, 2020
|
|
2G3A
 
 | | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens | | Descriptor: | acetyltransferase | | Authors: | Cymborowski, M, Xu, X, Chruszcz, M, Zheng, H, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens
To be Published
|
|
1HME
 
 | | STRUCTURE OF THE HMG BOX MOTIF IN THE B-DOMAIN OF HMG1 | | Descriptor: | HIGH MOBILITY GROUP PROTEIN FRAGMENT-B | | Authors: | Weir, H.M, Kraulis, P.J, Hill, C.S, Raine, A.R.C, Laue, E.D, Thomas, J.O. | | Deposit date: | 1994-02-10 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the HMG box motif in the B-domain of HMG1.
EMBO J., 12, 1993
|
|
4UUJ
 
 | | POTASSIUM CHANNEL KCSA-FAB WITH TETRAHEXYLAMMONIUM | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, COBALT (II) ION, ... | | Authors: | Lenaeus, M.J, Burdette, D, Wagner, T, Focia, P.J, Gross, A. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Kcsa in Complex with Symmetrical Quaternary Ammonium Compounds Reveal a Hydrophobic Binding Site.
Biochemistry, 53, 2014
|
|
8RM7
 
 | | Crystal Structure of Human Androgen Receptor DNA Binding Domain Bound to its Response Element: MMTV-177 GRE/ARE | | Descriptor: | Isoform 2 of Androgen receptor, MMTV-177 GRE/ARE Chain C, MMTV-177 GRE/ARE, ... | | Authors: | Lee, X.Y, Helsen, C, Van Eynde, W, Voet, A, Claessens, F. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural mechanism underlying variations in DNA binding by the androgen receptor.
J.Steroid Biochem.Mol.Biol., 241, 2024
|
|
8RXR
 
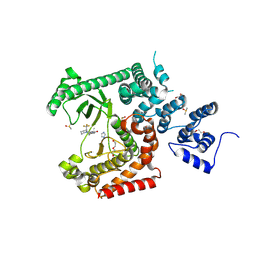 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | Descriptor: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 18, 2024
|
|
2G2D
 
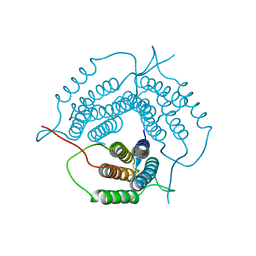 | | Crystal structure of a putative pduO-type ATP:cobalamin adenosyltransferase from Mycobacterium tuberculosis | | Descriptor: | ATP:cobalamin adenosyltransferase | | Authors: | Moon, J.H, Kaviratne, A, Yu, M, Bursey, E.H, Hung, L.-W, Lekin, T.P, Segelke, B.W, Terwilliger, T.C, Kim, C.-Y, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-02-15 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative pduO-type ATP:cobalamin adenosyltransferase from Mycobacterium tuberculosis.
To be Published
|
|
7S4Y
 
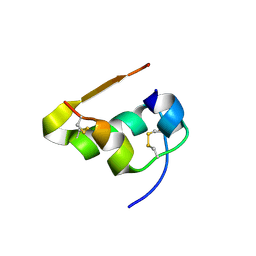 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
8HH8
 
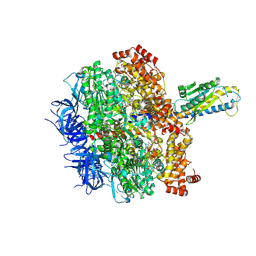 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd,lowATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8SIO
 
 | | Crystal structure of PRMT3 with YD1-66 | | Descriptor: | 5'-S-{3-[N'-(4'-chloro[1,1'-biphenyl]-3-yl)carbamimidamido]propyl}-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PRMT3 with YD1-66
To be published
|
|
4WID
 
 | | Crystal structure of the immediate-early 1 protein (IE1) at 2.31 angstrom (tetragonal form after crystal dehydration) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RhUL123 | | Authors: | Klingl, S, Scherer, M, Sevvana, M, Muller, Y.A, Stamminger, T. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions.
Plos Pathog., 10, 2014
|
|
2G5L
 
 | | Streptavidin in complex with Nanotag | | Descriptor: | (FME)(ASP)(VAL)(GLU)(ALA)(TRP)(LEU), GLYCEROL, SULFATE ION, ... | | Authors: | Perbandt, M, Bruns, O, Vallazza, M, Lamla, T, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-02-23 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High resolution structure of Streptavidin in complex with a novel high affinity peptide tag mimicking the biotin binding motif
To be Published
|
|
2G7J
 
 | | Solution NMR structure of the putative cytoplasmic protein ygaC from Salmonella typhimurium. Northeast Structural Genomics target StR72. | | Descriptor: | putative cytoplasmic protein | | Authors: | Aramini, J.M, Swapna, G.V.T, Ramelot, T.A, Ho, C.K, Cunningham, K, Ma, L.-C, Shetty, K, Xiao, R, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the putative cytoplasmic protein ygaC from Salmonella typhimurium. Northeast Structural Genomics target StR72.
To be Published
|
|
6DY8
 
 | |
7S89
 
 | | Open apo-state cryo-EM structure of human TRPV6 in cNW11 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
7S8C
 
 | | Cryo-EM structure of human TRPV6 in complex with inhibitor econazole | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
6DYJ
 
 | |
7S88
 
 | | Open apo-state cryo-EM structure of human TRPV6 in glyco-diosgenin detergent | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
4WLJ
 
 | | High resolution crystal structure of human kynurenine aminotransferase-I in complex with aminooxyacetate | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | High resolution crystal structures of human kynurenine aminotransferase-I bound to PLP cofactor, and in complex with aminooxyacetate.
Protein Sci., 26, 2017
|
|
7S8B
 
 | | Cryo-EM structure of human TRPV6 in complex with channel blocker ruthenium red | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
4WSC
 
 | | Crystal structure of a GroELK105A mutant | | Descriptor: | 60 kDa chaperonin | | Authors: | Lorimer, G.H, Ye, X, Fei, X, Yang, D, Corsepius, N, LaRonde, N.A. | | Deposit date: | 2014-10-26 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of a GroELK105A mutant
To Be Published
|
|
7RUA
 
 | | Metazoan pre-targeting GET complex (cBUGG-out) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 4 homolog, ... | | Authors: | Keszei, A.F.A, Yip, M.C.J, Shao, S. | | Deposit date: | 2021-08-16 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into metazoan pretargeting GET complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
