5U2K
 
 | | Crystal structure of Galactoside O-acetyltransferase complex with CoA (H3 space group) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Czub, M.P, Porebski, P.J, Knapik, A.A, Niedzialkowska, E, Siuda, M.K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-30 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of Galactoside O-acetyltransferase complex
with CoA (H3 space group)
to be published
|
|
3H4K
 
 | | Crystal structure of the wild type Thioredoxin glutatione reductase from Schistosoma mansoni in complex with auranofin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GOLD ION, ... | | Authors: | Angelucci, F, Dimastrogiovanni, D, Miele, A.E, Boumis, G, Brunori, M, Bellelli, A. | | Deposit date: | 2009-04-20 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Inhibition of Schistosoma mansoni thioredoxin-glutathione reductase by auranofin: structural and kinetic aspects.
J.Biol.Chem., 284, 2009
|
|
8GJ1
 
 | |
3H0Z
 
 | | Aurora A in complex with a bisanilinopyrimidine | | Descriptor: | 4-{[2-({4-[2-(4-acetylpiperazin-1-yl)-2-oxoethyl]phenyl}amino)-5-fluoropyrimidin-4-yl]amino}-N-(2-chlorophenyl)benzamide, Serine/threonine-protein kinase 6 | | Authors: | Wiesmann, C, Ultsch, M.H, Cochran, A.G. | | Deposit date: | 2009-04-10 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | A class of 2,4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B.
J.Med.Chem., 52, 2009
|
|
5GHL
 
 | | Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger | | Descriptor: | GLYCEROL, Glucoamylase, SULFATE ION | | Authors: | Miyake, H, Suyama, Y, Muraki, N, Kusunoki, M, Tanaka, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the starch-binding domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
6POR
 
 | | Antimicrobial lasso peptide ubonodin | | Descriptor: | Ubonodin | | Authors: | Link, A.J, Cheung-Lee, W.L. | | Deposit date: | 2019-07-05 | | Release date: | 2019-12-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of Ubonodin, an Antimicrobial Lasso Peptide Active against Members of the Burkholderia cepacia Complex.
Chembiochem, 21, 2020
|
|
5Y87
 
 | |
5U3V
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 6 | | Descriptor: | 6-[2-({cyclopentyl[4-(furan-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7L4I
 
 | |
6CL2
 
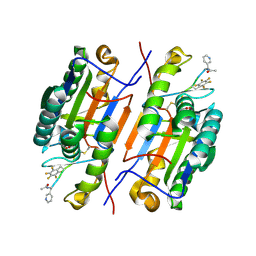 | |
8GJ2
 
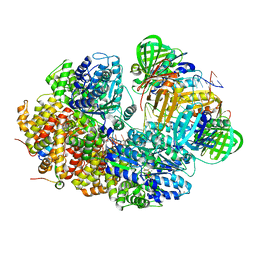 | |
6CLD
 
 | | 1.01 A MicroED structure of GSNQNNF at 0.81 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLK
 
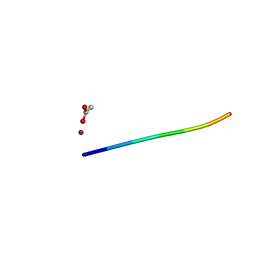 | | 1.01 A MicroED structure of GSNQNNF at 0.82 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
4QFZ
 
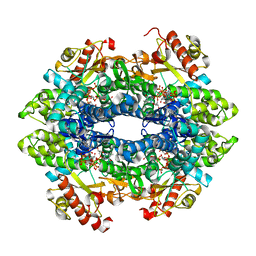 | | Crystal structure of the tetrameric dGTP/dTTP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
6CLS
 
 | | 1.46 A MicroED structure of GSNQNNF at 3.4 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.46 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
8GJ0
 
 | |
4YFX
 
 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
1AKZ
 
 | | HUMAN URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Tainer, J.A, Mol, C.D. | | Deposit date: | 1997-05-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
Embo J., 17, 1998
|
|
7BCP
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with gluconate | | Descriptor: | D-gluconic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
5MY2
 
 | |
6SQ9
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 3-hydroxynaphthalene-2-carboxylic acid | | Descriptor: | 3-hydroxynaphthalene-2-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6IXI
 
 | | structure of Cd-bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, CADMIUM ION, GLYCEROL, ... | | Authors: | Kumar, P, Sharma, N, Dalal, V, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-12-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of the heavy metal binding properties of periplasmic metal uptake protein CLas-ZnuA2.
Metallomics, 12, 2020
|
|
4YLE
 
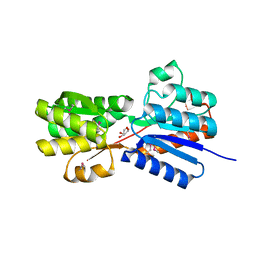 | | Crystal structure of an ABC transpoter solute binding protein (IPR025997) from Burkholderia multivorans (Bmul_1631, Target EFI-511115) with an unknown ligand modelled as alpha-D-erythrofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, UNKNOWN LIGAND | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-15 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Burkholderia multivorans (Bmul_1631, Target EFI-511115) with an unknown ligand modelled as alpha-D-erythrofuranose
To be published
|
|
8VBM
 
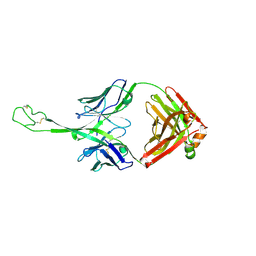 | | Structure of bovine anti-HIV Fab ElsE7 | | Descriptor: | Bovine Fab ElsE7 heavy chain, Bovine Fab ElsE7 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultra-long CDRH3 repertoire
To Be Published
|
|
4QJ8
 
 | |
