6HNF
 
 | | Structure in solution of human fibronectin type III-domain 14 | | 分子名称: | Fibronectin | | 著者 | Zhong, X, Arnolds, O, Krenczyk, O, Gajewski, J, Puetz, S, Herrmann, C, Stoll, R. | | 登録日 | 2018-09-14 | | 公開日 | 2018-10-10 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structure in Solution of Fibronectin Type III Domain 14 Reveals Its Synergistic Heparin Binding Site.
Biochemistry, 57, 2018
|
|
3CQ8
 
 | | Ternary complex of the L415F mutant RB69 exo(-)polymerase | | 分子名称: | CALCIUM ION, DNA (5'-D(*DAP*DCP*DAP*DGP*DGP*DTP*DAP*DAP*DGP*DCP*DAP*DGP*DTP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(*DGP*DCP*DGP*DGP*DAP*DCP*DTP*DGP*DCP*DTP*DTP*DAP*DCP*DC)-3'), ... | | 著者 | Zhong, X, Pedersen, L.C, Kunkel, T.A. | | 登録日 | 2008-04-02 | | 公開日 | 2008-09-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Characterization of a replicative DNA polymerase mutant with reduced fidelity and increased translesion synthesis capacity.
Nucleic Acids Res., 36, 2008
|
|
8GTJ
 
 | |
8GTK
 
 | |
1HUO
 
 | | CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP | | 分子名称: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*C)-3', CHROMIUM ION, ... | | 著者 | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | 登録日 | 2001-01-04 | | 公開日 | 2001-04-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
1HUZ
 
 | | CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP | | 分子名称: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*CP*T)-3', CHROMIUM ION, ... | | 著者 | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | 登録日 | 2001-01-04 | | 公開日 | 2001-04-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
4QD2
 
 | | Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex | | 分子名称: | CALCIUM ION, Cadherin-1, Hemagglutinin component HA17, ... | | 著者 | Lee, K, Zhong, X, Gu, S, Kruel, A, Dorner, M.B, Perry, K, Rummel, A, Dong, M, Jin, R. | | 登録日 | 2014-05-13 | | 公開日 | 2014-06-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex.
Science, 344, 2014
|
|
6VII
 
 | |
6VIK
 
 | |
6VIH
 
 | |
6VIJ
 
 | |
4ONJ
 
 | |
4ONQ
 
 | |
8GTN
 
 | |
8TN1
 
 | |
8TN6
 
 | |
8TNB
 
 | |
8TNC
 
 | |
8TND
 
 | |
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
4E3C
 
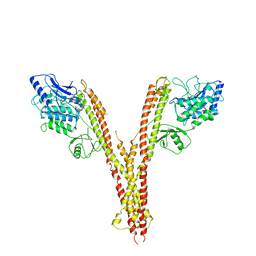 | | X-ray crystal structure of human IKK2 in an active conformation | | 分子名称: | Inhibitor of nuclear factor kappa-B kinase subunit beta | | 著者 | Polley, S, Huang, D.B, Hauenstein, A.V, Ghosh, G, Huxford, T. | | 登録日 | 2012-03-09 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.98 Å) | | 主引用文献 | X-ray crystal structure of human IKK2 in an active conformation
Plos.Biol., 2013
|
|
2IHM
 
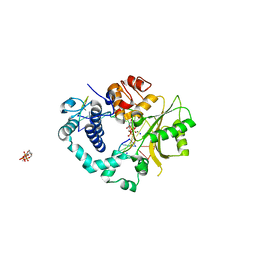 | | Polymerase mu in ternary complex with gapped 11mer DNA duplex and bound incoming nucleotide | | 分子名称: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*T)-3', 5'-D(*CP*GP*GP*CP*AP*AP*TP*AP*CP*TP*G)-3', ... | | 著者 | Moon, A.F, Pedersen, L.C, Kunkel, T.A. | | 登録日 | 2006-09-26 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insight into the substrate specificity of DNA Polymerase mu.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2ID5
 
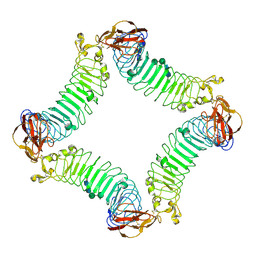 | | Crystal Structure of the Lingo-1 Ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine rich repeat neuronal 6A, ... | | 著者 | Mosyak, L, Wood, A, Dwyer, B, Johnson, M, Stahl, M.L, Somers, W.S. | | 登録日 | 2006-09-14 | | 公開日 | 2006-09-26 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.698 Å) | | 主引用文献 | The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition.
J.Biol.Chem., 281, 2006
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
