8ZC5
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike
To be published
|
|
5WSH
 
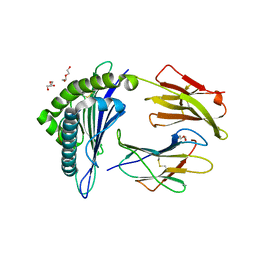 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLY-VAL-TRP-ILE-ARG-THR-PRO-THR-ALA, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2016-12-07 | | Release date: | 2017-12-20 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
8C3W
 
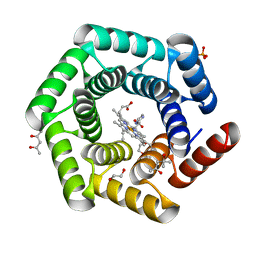 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
6LPF
 
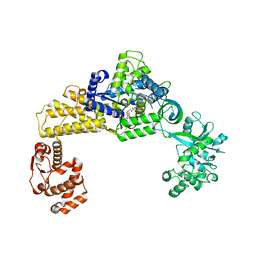 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-10 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
6LR6
 
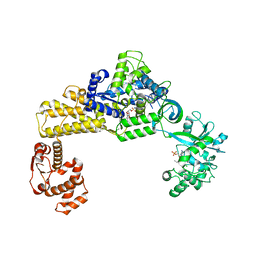 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
3H59
 
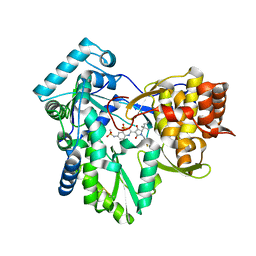 | | Hepatitis C virus polymerase NS5B with thiazine inhibitor 2 | | Descriptor: | N-{3-[(5S)-5-(1,1-dimethylpropyl)-1-(4-fluoro-3-methylbenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-4H-1,4-benzothiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-04-21 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 3: synthesis and optimization studies of benzothiazine-substituted tetramic acids
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1BKO
 
 | |
1BSF
 
 | |
1BKP
 
 | |
1BSP
 
 | | THERMOSTABLE THYMIDYLATE SYNTHASE A FROM BACILLUS SUBTILIS | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE A | | Authors: | Stout, T.J, Schellenberger, U, Santi, D.V, Stroud, R.M. | | Deposit date: | 1998-07-09 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of a unique thermal-stable thymidylate synthase from Bacillus subtilis.
Biochemistry, 37, 1998
|
|
8IYX
 
 | | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | | Descriptor: | 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid, Probable G-protein coupled receptor 34,YL-365 | | Authors: | Jia, G.W, Wang, X, Zhang, C.B, Dong, H.H, Su, Z.M. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6CEO
 
 | |
7DAV
 
 | | The native crystal structure of COVID-19 main protease | | Descriptor: | COVID-19 MAIN PROTEASE | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7DAT
 
 | | The crystal structure of COVID-19 main protease treated by AF | | Descriptor: | COVID-19 MAIN PROTEASE, GOLD ION | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7DAU
 
 | | The crystal structure of COVID-19 main protease treated by GA | | Descriptor: | COVID-19 MAIN PROTEASE, GOLD ION | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
1I7E
 
 | | C-Terminal Domain Of Mouse Brain Tubby Protein bound to Phosphatidylinositol 4,5-bis-phosphate | | Descriptor: | L-ALPHA-GLYCEROPHOSPHO-D-MYO-INOSITOL-4,5-BIS-PHOSPHATE, TUBBY PROTEIN | | Authors: | Santagata, S, Boggon, T.J, Baird, C.L, Shan, W.S, Shapiro, L. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | G-protein signaling through tubby proteins.
Science, 292, 2001
|
|
5VXA
 
 | | Structure of the human Mesh1-NADPH complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-05-23 | | Release date: | 2018-05-23 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MESH1 is a cytosolic NADPH phosphatase that regulates ferroptosis.
Nat Metab, 2, 2020
|
|
1R9H
 
 | |
5DRO
 
 | | Structure of the Aquifex aeolicus LpxC/LPC-011 Complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRP
 
 | | Structure of the AaLpxC/LPC-023 Complex | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N~2~-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-isoleucinamide, ... | | Authors: | Najeeb, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRR
 
 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-058 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
3H2L
 
 | | Crystal structure of HCV NS5B polymerase in complex with a novel bicyclic dihydro-pyridinone inhibitor | | Descriptor: | N-{3-[(4aR,7aS)-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-2,4a,5,6,7,7a-hexahydro-1H-cyclopenta[b]pyridin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, NS5B polymerase | | Authors: | Han, Q, Showalter, R.E, Zhou, Q, Kissinger, C.R. | | Deposit date: | 2009-04-14 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of tricyclic 5,6-dihydro-1H-pyridin-2-ones as novel, potent, and orally bioavailable inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5DRQ
 
 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-040 complex | | Descriptor: | N-[(2S)-3-amino-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
3CWJ
 
 | | Crystal structure of hcv ns5b polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,4-benzothiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-04-21 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 4-(1,1-Dioxo-1,4-dihydro-1lambda6-benzo[1,4]thiazin-3-yl)-5-hydroxy-2H-pyridazin-3-ones as potent inhibitors of HCV NS5B polymerase
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CO9
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[4-hydroxy-1-(3-methylbutyl)-2-oxo-1,2-dihydropyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7 -yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-03-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrrolo[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
