1YND
 
 | | Structure of human cyclophilin A in complex with the novel immunosuppressant sanglifehrin A at 1.6A resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, SANGLIFEHRIN A | | Authors: | Kallen, J, Sedrani, R, Zenke, G, Wagner, J. | | Deposit date: | 2005-01-24 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human cyclophilin A in complex with the novel immunosuppressant sanglifehrin A at 1.6 A resolution.
J.Biol.Chem., 280, 2005
|
|
3IW4
 
 | | Crystal structure of PKC alpha in complex with NVP-AEB071 | | Descriptor: | 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]-1H-pyrrole-2,5-dione, Protein kinase C alpha type | | Authors: | Stark, W, Rummel, G, Strauss, A, Cowan-Jacob, S.W. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]pyrrole-2,5-dione (AEB071), a potent and selective inhibitor of protein kinase C isotypes
J.Med.Chem., 52, 2009
|
|
1NMK
 
 | | The Sanglifehrin-Cyclophilin Interaction: Degradation Work, Synthetic Macrocyclic Analogues, X-ray Crystal Structure and Binding Data | | Descriptor: | (13E,15E)-(3S,6S,9R,10R,11S,12S,18S,21S)-10,12-DIHYDROXY-3-(3-HYDROXYBEN-ZYL)-18-((E)-3-HYDROXY-1-METHYLPROPENYL)-6-ISOPROPYL-11-METHYL-9-(3-OXO-BUTYL)-19-OXA-1,4,7,25-TETRAAZA-BICYCLO[19.3.1]PENTACOSA-13,15-DIENE-2,5,8,20-TETRAONE, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Kallen, J, Sedrani, R, Wagner, J. | | Deposit date: | 2003-01-10 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sanglifehrin-Cyclophilin Interaction: Degradation Work, Synthetic Macrocyclic Analogues, X-ray Crystal Structure and Binding Data
J.Am.Chem.Soc., 125, 2003
|
|
8RY2
 
 | |
5LQB
 
 | |
6GGS
 
 | | Structure of RIP2 CARD filament | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pellegrini, E, Cusack, S, Desfosses, A, Schoehn, G, Malet, H, Gutsche, I, Sachse, C, Hons, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
6GFJ
 
 | | Structure of RIP2 CARD domain fused to crystallisable MBP tag | | Descriptor: | Sugar ABC transporter substrate-binding protein,Receptor-interacting serine/threonine-protein kinase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2018-04-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
7OZT
 
 | | Nanobodies restore stability to cancer-associated mutants of tumor suppressor protein p16INK4a | | Descriptor: | Camelid nanobody NB09, Cyclin-dependent kinase inhibitor 2A | | Authors: | Pastok, M.W, Burbidge, O, Itzhaki, L, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Nanobodies restore stability to cancer-associated mutants of tumor suppressor protein p16INK4a
To Be Published
|
|
4RCL
 
 | |
5VBA
 
 | |
5DLB
 
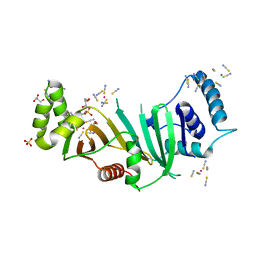 | | Crystal structure of chaperone EspG3 of ESX-3 type VII secretion system from Mycobacterium marinum M | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, PLATINUM (II) ION, ... | | Authors: | Chan, S, Arbing, M.A, Kim, J, Kahng, S, Sawaya, M.R, Eisenberg, D.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Variability of EspG Chaperones from Mycobacterial ESX-1, ESX-3, and ESX-5 Type VII Secretion Systems.
J. Mol. Biol., 431, 2019
|
|
4L4W
 
 | |
5SXL
 
 | |
