7MT0
 
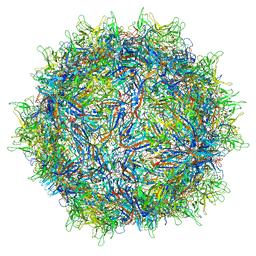 | | Structure of the adeno-associated virus 9 capsid at pH 7.4 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTZ
 
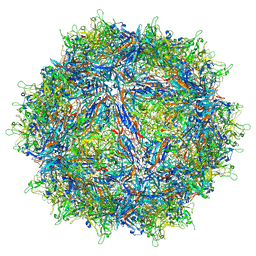 | | Structure of the adeno-associated virus 9 capsid at pH pH 7.4 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MUA
 
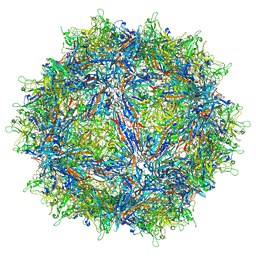 | | Structure of the adeno-associated virus 9 capsid at pH pH 5.5 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTG
 
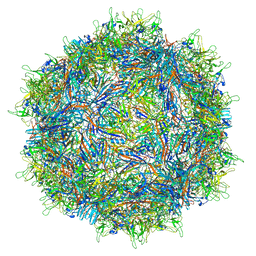 | | Structure of the adeno-associated virus 9 capsid at pH 6.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTP
 
 | | Structure of the adeno-associated virus 9 capsid at pH 5.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTW
 
 | | Structure of the adeno-associated virus 9 capsid at pH 4.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
8TEX
 
 | | Avian Adeno-associated virus - empty capsid | | Descriptor: | Capsid protein | | Authors: | Hsi, J, Mietzsch, M, Chipman, P, Afione, S, Zeher, A, Huang, R, Chiorini, J, McKenna, R. | | Deposit date: | 2023-07-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural and antigenic characterization of the avian adeno-associated virus capsid.
J.Virol., 97, 2023
|
|
8TEY
 
 | | Avian Adeno-associated virus - empty capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Hsi, J, Mietzsch, M, Chipman, P, Afione, S, Zeher, A, Huang, R, Chiorini, J, McKenna, R. | | Deposit date: | 2023-07-07 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and antigenic characterization of the avian adeno-associated virus capsid.
J.Virol., 97, 2023
|
|
7TZ6
 
 | | Structure of mitochondrial bc1 in complex with ck-2-68 | | Descriptor: | 7-chloranyl-3-methyl-2-[4-[[4-(trifluoromethyloxy)phenyl]methyl]phenyl]-1~{H}-quinolin-4-one, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Huang, R. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure of complex III with bound antimalarial agent CK-2-68 provides insights into selective inhibition of Plasmodium cytochrome bc 1 complexes.
J.Biol.Chem., 299, 2023
|
|
7NAB
 
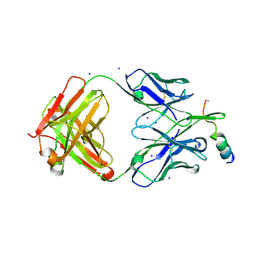 | | Crystal structure of human neutralizing mAb CV3-25 binding to SARS-CoV-2 S MPER peptide 1140-1165 | | Descriptor: | CITRIC ACID, CV3-25 Fab Heavy Chain, CV3-25 Fab Light Chain, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-06-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Cell Rep, 38, 2022
|
|
7N0H
 
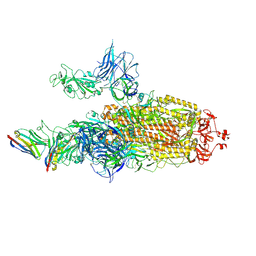 | | CryoEM structure of SARS-CoV-2 spike protein (S-6P, 2-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7N0G
 
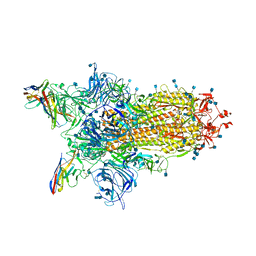 | | CryoEm structure of SARS-CoV-2 spike protein (S-6P, 1-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KLW
 
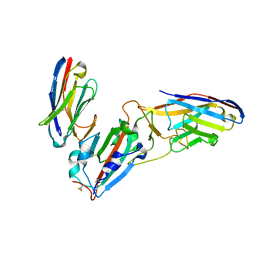 | | Crystal structure of synthetic nanobody (Sb45+Sb68) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | SB45, Synthetic Nanobody, SB68, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-11-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7MFU
 
 | | Crystal structure of synthetic nanobody (Sb14+Sb68) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Spike protein S1, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7MFV
 
 | | Crystal structure of synthetic nanobody (Sb16) | | Descriptor: | 1,2-ETHANEDIOL, Synthetic Nanobody #16 (Sb16) | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KGJ
 
 | | Crystal structure of synthetic nanobody (Sb45) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | Sb45, Sybody-45, Synthetic Nanobody, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-10-16 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KGK
 
 | | Crystal structure of synthetic nanobody (Sb16) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | Sb16, Sybody-16, Synthetic Nanobody, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-10-16 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
8T5U
 
 | | ATP-1 state of Bcs1 (C7 symmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-06-14 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8T7U
 
 | | ADP-bound Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8T14
 
 | | ADP-bound Bcs1 (C7 symmetrized) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TBY
 
 | | Apo Bcs1, unsymmetrized | | Descriptor: | Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-06-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TI0
 
 | | ATP-1 state of Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-07-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TPL
 
 | | ATP-2 state of Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TP1
 
 | | ATP-2 state of Bcs1 (C7 symmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
