2KHZ
 
 | | Solution Structure of RCL | | Descriptor: | c-Myc-responsive protein Rcl | | Authors: | Doddapaneni, K, Mahler, B, Yuan, C, Wu, Z. | | Deposit date: | 2009-04-15 | | Release date: | 2009-10-13 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RCL, a novel 2'-deoxyribonucleoside 5'-monophosphate N-glycosidase
J.Mol.Biol., 394, 2009
|
|
4H42
 
 | | Synthesis of a Weak Basic uPA Inhibitor and Crystal Structure of Complex with uPA | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, N-[(2-amino-1,3-benzothiazol-6-yl)carbonyl]glycine, Urokinase-type plasminogen activator | | Authors: | Yu, H.-Y, Gao, D, Zhang, X, Jiang, L.-G, Hong, Z.-B, Yuan, C, Fang, X, Wang, J.-D, Huang, M.-D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Synthesis of a Weak Basic uPA Inhibitor and Crystal Structure of Complex with uPA
CHIN.J.STRUCT.CHEM., 32, 2013
|
|
4ISO
 
 | | Crystal Structure of Matriptase in complex with its inhibitor HAI-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, GLYCEROL, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
4ISN
 
 | | Crystal Structure of Matriptase in complex with its inhibitor HAI-1 | | Descriptor: | GLUTATHIONE, Kunitz-type protease inhibitor 1, Suppressor of tumorigenicity 14 protein, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
4IS5
 
 | | Crystal Structure of the ligand-free inactive Matriptase | | Descriptor: | GLUTATHIONE, GLYCEROL, SULFATE ION, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
4ISL
 
 | | Crystal Structure of the inactive Matriptase in complex with its inhibitor HAI-1 | | Descriptor: | GLUTATHIONE, GLYCEROL, Kunitz-type protease inhibitor 1, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
3M61
 
 | | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1 W3A | | Authors: | Jiang, L, Yuan, C, Wind, T, Andreasen, P.A, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition
TO BE PUBLISHED
|
|
3MHW
 
 | | The complex crystal Structure of Urokianse and 2-Aminobenzothiazole | | Descriptor: | 1,3-benzothiazol-2-amine, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.-G, Yuan, C, Chen, L.-Q, Huang, M.-D. | | Deposit date: | 2010-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
3MWI
 
 | | The complex crystal Structure of Urokianse and 5-nitro-1H-indole-2-amidine | | Descriptor: | 5-nitro-1H-indole-2-carboximidamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Yu, H.Y, Yuan, C, Huang, Z.X, Huang, M.D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The complex crystal Structure of Urokianse and 5-nitro-1H-indole-2-amidine
To be Published
|
|
3U74
 
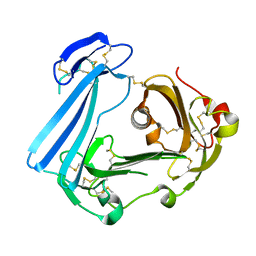 | | Crystal structure of stabilized human uPAR mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Huang, M.D, Xu, X, Yuan, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the urokinase receptor in a ligand-free form.
J.Mol.Biol., 416, 2012
|
|
3U73
 
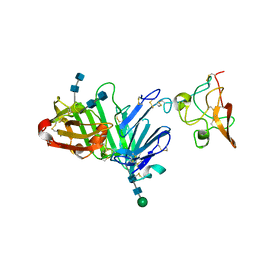 | | Crystal structure of stabilized human uPAR mutant in complex with ATF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, Urokinase-type plasminogen activator, ... | | Authors: | Huang, M.D, Xu, X, Yuan, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-04-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of the urokinase receptor in a ligand-free form.
J.Mol.Biol., 416, 2012
|
|
7F6L
 
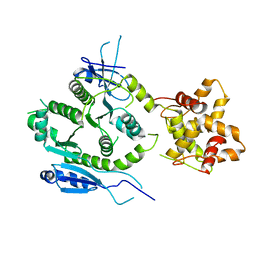 | | Crystal structure of human MUS81-EME2 complex | | Descriptor: | Crossover junction endonuclease MUS81, Probable crossover junction endonuclease EME2 | | Authors: | Hua, Z.K, Zhang, D.P, Yuan, C, Lin, Z.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human MUS81-EME2 complex.
Structure, 30, 2022
|
|
7WOK
 
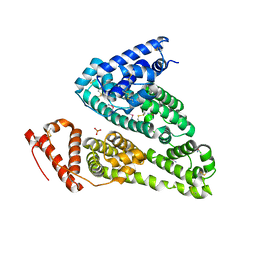 | | Crystal structure of HSA soaked with cisplatin for one week | | Descriptor: | Albumin, Cisplatin, PHOSPHATE ION | | Authors: | Chen, S.L, Yuan, C, Jiang, L.G, Luo, Z.P, Huang, M.D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic analysis of interaction between cisplatin and human serum albumin: Effect of fatty acid.
Int.J.Biol.Macromol., 216, 2022
|
|
7WOJ
 
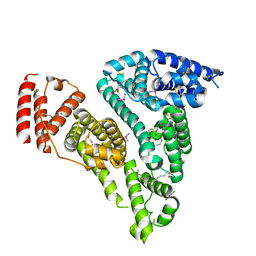 | | Crystal structure of HSA-Myr complex soaked with cisplatin for one week | | Descriptor: | Albumin, Cisplatin, MYRISTIC ACID | | Authors: | Chen, S.L, Yuan, C, Jiang, L.G, Luo, Z.P, Huang, M.D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystallographic analysis of interaction between cisplatin and human serum albumin: Effect of fatty acid.
Int.J.Biol.Macromol., 216, 2022
|
|
6IS8
 
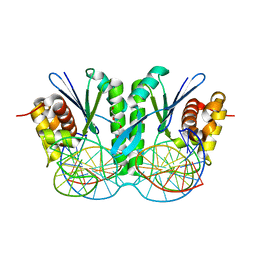 | | Crystal structure of ZmMoc1 D115N mutant in complex with Holliday junction | | Descriptor: | DNA (33-MER), MAGNESIUM ION, Monokaryotic chloroplast 1, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
6IS9
 
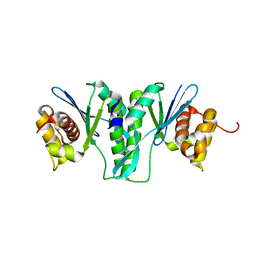 | | Crystal Structure of ZmMOC1 | | Descriptor: | Monokaryotic chloroplast 1 | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
6ION
 
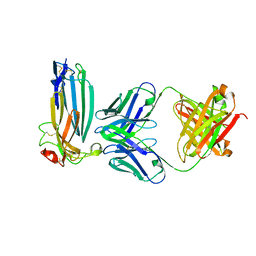 | | The complex of C4.4A with its antibody 11H10 Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ly6/PLAUR domain-containing protein 3, ... | | Authors: | Huang, M.D, Jiang, Y.B, Yuan, C, Lin, L. | | Deposit date: | 2018-10-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Human C4.4A Reveal the Unique Association of Ly6/uPAR/alpha-neurotoxin Domain
Int J Biol Sci, 16, 2020
|
|
6IOM
 
 | | Crystal structure of human C4.4A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ly6/PLAUR domain-containing protein 3 | | Authors: | Huang, M.D, Jiang, Y.B, Yuan, C, Lin, L. | | Deposit date: | 2018-10-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Crystal Structures of Human C4.4A Reveal the Unique Association of Ly6/uPAR/alpha-neurotoxin Domain
Int J Biol Sci, 16, 2020
|
|
6JRG
 
 | | Crystal structure of ZmMoc1 H253A mutant in complex with Holliday junction | | Descriptor: | DNA (32-MER), DNA (33-MER), MAGNESIUM ION, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
6JRF
 
 | | Crystal structure of ZmMoc1-Holliday junction Complex in the presence of Calcium | | Descriptor: | CALCIUM ION, DNA (33-MER), Monokaryotic chloroplast 1, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
1BVM
 
 | | SOLUTION NMR STRUCTURE OF BOVINE PANCREATIC PHOSPHOLIPASE A2, 20 STRUCTURES | | Descriptor: | PROTEIN (PHOSPHOLIPASE A2) | | Authors: | Yuan, C.-H, Byeon, I.-J.L, Li, Y, Tsai, M.-D. | | Deposit date: | 1998-09-14 | | Release date: | 1999-09-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of phospholipase A2 from functional perspective. 1. Functionally relevant solution structure and roles of the hydrogen-bonding network.
Biochemistry, 38, 1999
|
|
2RBE
 
 | | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors | | Descriptor: | (5R)-2-[(2-fluorophenyl)amino]-5-(1-methylethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Jordan, S.R, Li, V. | | Deposit date: | 2007-09-18 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8HUZ
 
 | |
4K23
 
 | | Structure of anti-uPAR Fab ATN-658 | | Descriptor: | anti-uPAR antibody, heavy chain, light chain | | Authors: | Yuan, C, Huang, M, Chen, L. | | Deposit date: | 2013-04-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a New Epitope in uPAR as a Target for the Cancer Therapeutic Monoclonal Antibody ATN-658, a Structural Homolog of the uPAR Binding Integrin CD11b ( alpha M)
Plos One, 9, 2014
|
|
6XP5
 
 | | Head-Middle module of Mediator | | Descriptor: | HEAT, Med22, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Zhang, H.Q, Chen, D.C, Kornberg, R.D. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
