8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
7XTQ
 
 | | Cryo-EM structure of the R399-bound GPBAR-Gs complex | | Descriptor: | G-protein coupled bile acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, L, Yang, F, Wu, X, Mao, C, Sun, J, Yu, X, Zhang, Y, Zhang, P. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis and molecular mechanism of biased GPBAR signaling in regulating NSCLC cell growth via YAP activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5WDA
 
 | | Structure of the PulG pseudopilus | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Thomassin, J.L, Bardiaux, B, Zheng, W, Nivaskumar, M, Yu, X, Nilges, M, Egelman, E.H, Izadi-Pruneyre, N, Francetic, O. | | Deposit date: | 2017-07-04 | | Release date: | 2017-10-25 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
6BQ1
 
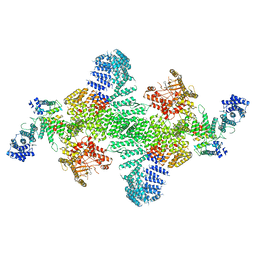 | | Human PI4KIIIa lipid kinase complex | | Descriptor: | 5-{2-amino-1-[4-(morpholin-4-yl)phenyl]-1H-benzimidazol-6-yl}-N-(2-fluorophenyl)-2-methoxypyridine-3-sulfonamide, Phosphatidylinositol 4-kinase III alpha (PI4KA), Protein FAM126A, ... | | Authors: | Lees, J.A, Zhang, Y, Oh, M, Schauder, C.M, Yu, X, Baskin, J, Dobbs, K, Notarangelo, L.D, Camilli, P.D, Walz, T, Reinisch, K.M. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the human PI4KIII alpha lipid kinase complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7WUJ
 
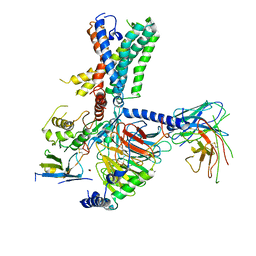 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G4,Uncharacterized protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Guo, S.C, Huang, S.M, He, Q.T, Xiao, P, Sun, J.P, Yu, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
7WUI
 
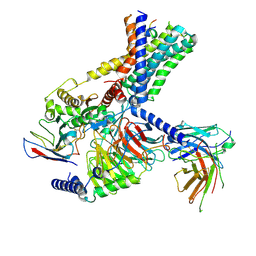 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Guo, S.C, He, Q.T, Xiao, P, Sun, J.P, Yu, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
7WUQ
 
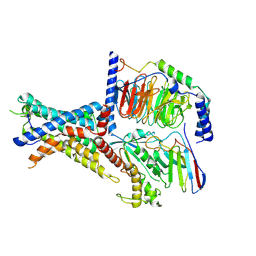 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Q.T, Guo, S.C, Xiao, P, Sun, J.P, Yu, X, Gou, L, Kong, L.L, Zhang, L. | | Deposit date: | 2022-02-09 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
8HEV
 
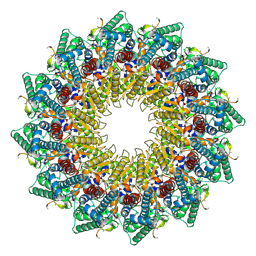 | | C12 portal in HCMV B-capsid | | Descriptor: | Portal protein, Unknown peptide | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HEU
 
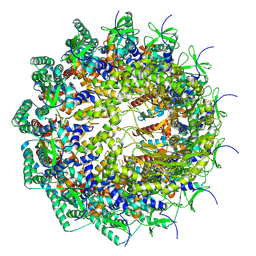 | | C12 portal in HCMV A-capsid | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HQE
 
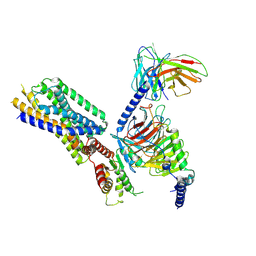 | | Cryo-EM structure of the apo-GPR132-Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HVI
 
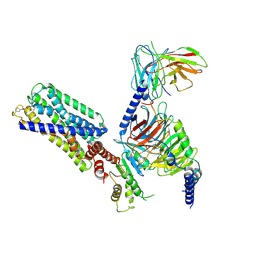 | | Activation mechanism of GPR132 by compound NOX-6-7 | | Descriptor: | 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofuran-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-26 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQN
 
 | | Activation mechanism of GPR132 by 9(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQM
 
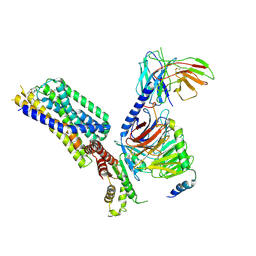 | | Activation mechanism of GPR132 by NPGLY | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HEX
 
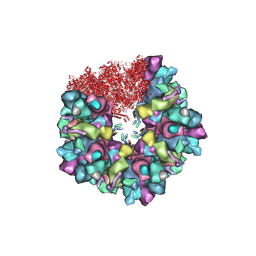 | | C5 portal vertex in HCMV B-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HEY
 
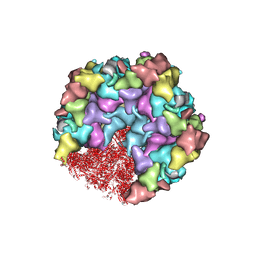 | | One CVSC-binding penton vertex in HCMV B-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
7DFB
 
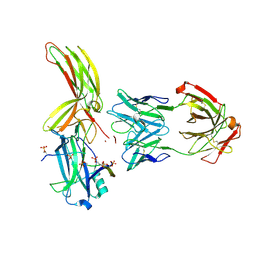 | | Crystal of Arrestin2-V2Rpp-6-7-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DF9
 
 | | Crystal of Arrestin2-V2Rpp-1-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFA
 
 | | Crystal of Arrestin2-V2Rpp-4-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFC
 
 | | Crystal of Arrestin2-V2Rpp-3-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7ETM
 
 | | C6 portal vertex in the enveloped virion capsid | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7ETO
 
 | | C1 CVSC-binding penton vertex in the virion capsid of Human Cytomegalovirus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7ETJ
 
 | | C5 portal vertex in the partially-enveloped virion capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7ET2
 
 | | Human Cytomegalovirus, C12 portal | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
