7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
8Z1P
 
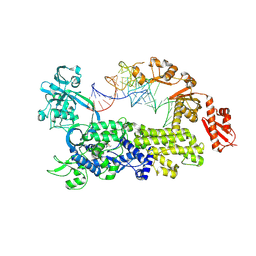 | | Crystal structure of Saccharomyces cerevisiae isoleucyl-tRNA synthetase in complex with a mimic tRNA(Met) and isoleucine | | Descriptor: | (2S,3S)-2-amino-3-methylpentane-1,1-diol, isoleucine--tRNA ligase, tRNA(Met) | | Authors: | Chen, B, Yi, F, Zhou, H. | | Deposit date: | 2024-04-11 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The mechanism of discriminative aminoacylation by isoleucyl-tRNA synthetase based on wobble nucleotide recognition.
Nat Commun, 15, 2024
|
|
7D77
 
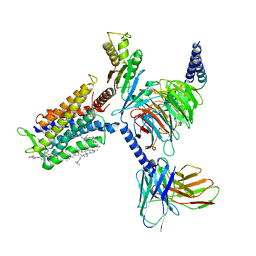 | | Cryo-EM structure of the cortisol-bound adhesion receptor GPR97-Go complex | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | Descriptor: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
7CFM
 
 | | Cryo-EM structure of the P395-bound GPBAR-Gs complex | | Descriptor: | 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
8WND
 
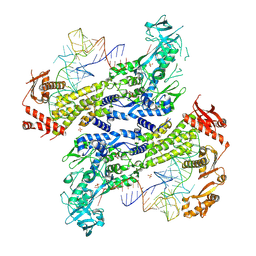 | | Crystal structure of Saccharomyces cerevisiae isoleucyl-tRNA synthetase in complex with tRNA(Ile) and isoleucine | | Descriptor: | 1,2-ETHANEDIOL, ISOLEUCINE, SULFATE ION, ... | | Authors: | Chen, B, Yi, F, Zhou, H. | | Deposit date: | 2023-10-05 | | Release date: | 2024-10-09 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mechanism of discriminative aminoacylation by isoleucyl-tRNA synthetase based on wobble nucleotide recognition.
Nat Commun, 15, 2024
|
|
5HPY
 
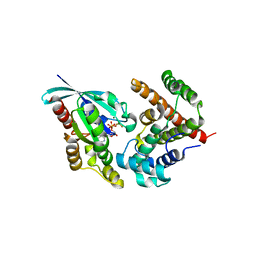 | | Crystal Structure of RhoA.GDP.MgF3-in complex with human Myosin 9b RhoGAP domain | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, TRIFLUOROMAGNESATE, ... | | Authors: | Yi, F.S, Ren, J.Q, Feng, W. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Noncanonical Myo9b-RhoGAP Accelerates RhoA GTP Hydrolysis by a Dual-Arginine-Finger Mechanism
J.Mol.Biol., 428, 2016
|
|
5I56
 
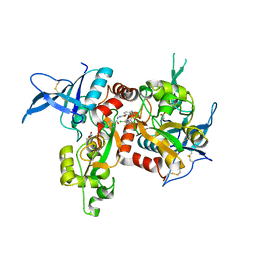 | | Agonist-bound GluN1/GluN2A agonist binding domains with TCN201 | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
5JTY
 
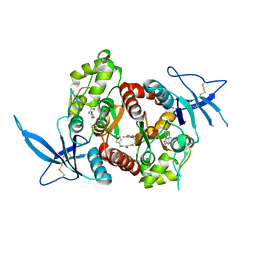 | | Glutamate- and DCKA-bound GluN1/GluN2A agonist binding domains with MPX-007 | | Descriptor: | 4-hydroxy-5,7-dimethylquinoline-2-carboxylic acid, 5-({[(3,4-difluorophenyl)sulfonyl]amino}methyl)-6-methyl-N-[(2-methyl-1,3-thiazol-5-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-05-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
5I59
 
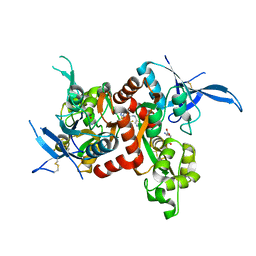 | | Glutamate- and glycine-bound GluN1/GluN2A agonist binding domains with MPX 007 | | Descriptor: | 5-({[(3,4-difluorophenyl)sulfonyl]amino}methyl)-6-methyl-N-[(2-methyl-4H-1lambda~4~,3-thiazol-5-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
5I58
 
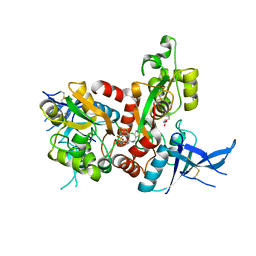 | | GLUTAMATE- AND GLYCINE-BOUND GLUN1/GLUN2A AGONIST BINDING DOMAINS WITH MPX-004 | | Descriptor: | 5-({[(3-chloro-4-fluorophenyl)sulfonyl]amino}methyl)-N-[(2-methyl-1,3-thiazol-5-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
5I57
 
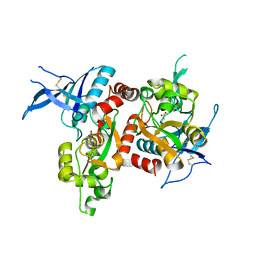 | | Glutamate- and glycine-bound GluN1/GluN2A agonist binding domains | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
8Y62
 
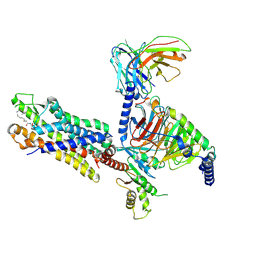 | | Cryo-EM structure of the C16:0 ceramide-bound FPR2-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Sun, J.P, Jiang, C.T, Kong, W, Yu, X, Cai, K, Guo, L.L. | | Deposit date: | 2024-02-01 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Metabolic signaling of ceramides through the FPR2 receptor inhibits adipocyte thermogenesis.
Science, 2025
|
|
8Y63
 
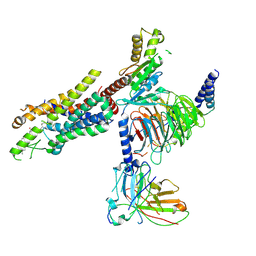 | | Cryo-EM structure of the C20:0 ceramide-bound FPR2-Gi complex | | Descriptor: | Cer(d18:0/20:0), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Jiang, C.T, Kong, W, Yu, X, Cai, K, Guo, L.L. | | Deposit date: | 2024-02-01 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Metabolic signaling of ceramides through the FPR2 receptor inhibits adipocyte thermogenesis.
Science, 2025
|
|
5C5S
 
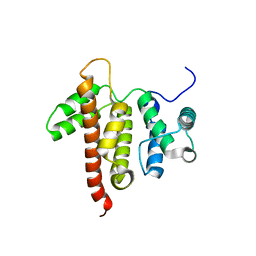 | |
7RRG
 
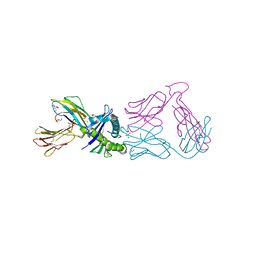 | |
5J8R
 
 | | Crystal Structure of the Catalytic Domain of Human Protein Tyrosine Phosphatase non-receptor Type 12 - K61R mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Li, H, Yang, F, Xu, Y.F, Wang, W.J, Xiao, P, Yu, X, Sun, J.P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Crystal structure and substrate specificity of PTPN12.
Cell Rep, 15, 2016
|
|
8ZFD
 
 | |
8ZD1
 
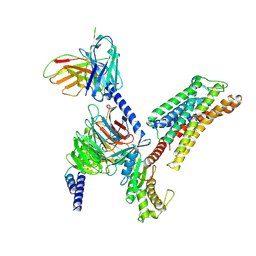 | | Cryo-EM structure of the xGPR4-Gs complex in pH6.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | Deposit date: | 2024-04-30 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZF6
 
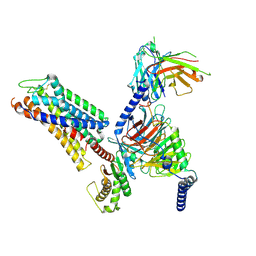 | | Cryo-EM structure of the xGPR4-Gs complex in pH6.7 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZF7
 
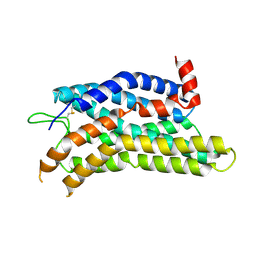 | |
8ZFA
 
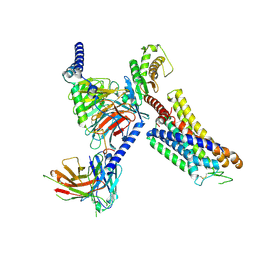 | | Cryo-EM structure of the xtGPR4-Gs complex in pH7.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZFE
 
 | |
8ZF4
 
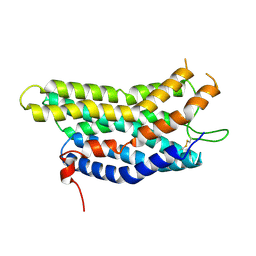 | |
8ZF9
 
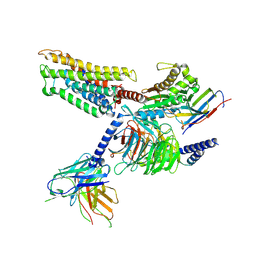 | | Cryo-EM structure of the mmGPR4-Gs complex in pH7.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wen, X, Rong, N.K, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
