3L43
 
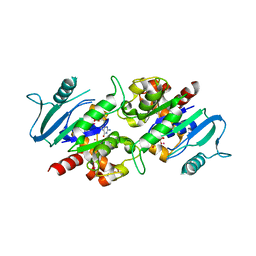 | | Crystal structure of the dynamin 3 GTPase domain bound with GDP | | 分子名称: | Dynamin-3, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | 著者 | Yang, S, Tempel, W, Tong, Y, Nedyalkova, L, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2009-12-18 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystal structure of the dynamin 3 GTPase domain bound with GDP
to be published
|
|
6VNW
 
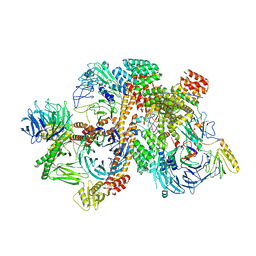 | | Cryo-EM structure of apo-BBSome | | 分子名称: | BBS1 domain-containing protein, Bardet-Biedl syndrome 18 protein, Bardet-Biedl syndrome 2 protein homolog, ... | | 著者 | Yang, S, Walz, T, Nachury, M, Chou, H. | | 登録日 | 2020-01-29 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes.
Elife, 9, 2020
|
|
6VOA
 
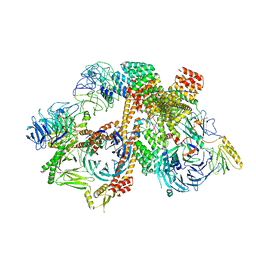 | | Cryo-EM structure of the BBSome-ARL6 complex | | 分子名称: | ADP-ribosylation factor-like protein 6, BBS1 domain-containing protein, Bardet-Biedl syndrome 18 protein, ... | | 著者 | Yang, S, Walz, T, Nachury, M.V. | | 登録日 | 2020-01-30 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes.
Elife, 9, 2020
|
|
2KAP
 
 | | Solution structure of DLC1-SAM | | 分子名称: | Rho GTPase-activating protein 7 | | 著者 | Yang, S, Yang, D. | | 登録日 | 2008-11-12 | | 公開日 | 2009-10-20 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Characterization of DLC1-SAM equilibrium unfolding at the amino acid residue level
Biochemistry, 48, 2009
|
|
5JJY
 
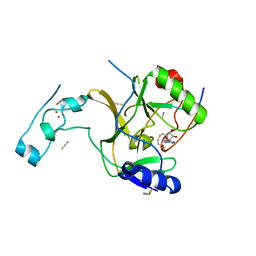 | | Crystal structure of SETD2 bound to histone H3.3 K36M peptide | | 分子名称: | Histone H3.3, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Yang, S, Zheng, X, Li, H. | | 登録日 | 2016-04-25 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.053 Å) | | 主引用文献 | Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase
Genes Dev., 30, 2016
|
|
6BD4
 
 | | Crystal structure of human apo-Frizzled4 receptor | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Frizzled-4/Rubredoxin chimeric protein, OLEIC ACID, ... | | 著者 | Yang, S, Wu, Y, Pu, M, Chen, Y, Dong, S, Guo, Y, Han, G.Y, Stevens, R.C, Zhao, S, Xu, F. | | 登録日 | 2017-10-21 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the Frizzled 4 receptor in a ligand-free state.
Nature, 560, 2018
|
|
2KET
 
 | |
7SC5
 
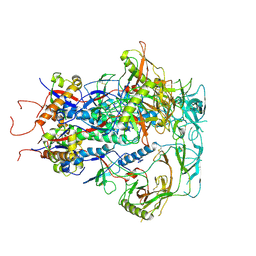 | | Cytoplasmic tail deleted HIV Env trimer in nanodisc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Yang, S, Walz, T. | | 登録日 | 2021-09-27 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
7SD3
 
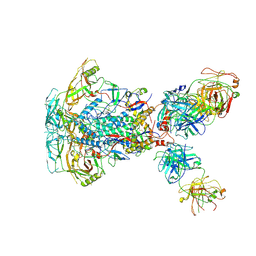 | | Cytoplasmic tail deleted HIV-1 Env bound with three 4E10 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4E10 Fab heavy chain, ... | | 著者 | Yang, S, Walz, T. | | 登録日 | 2021-09-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
5XBZ
 
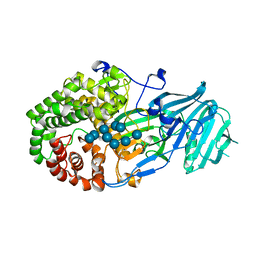 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminaripentaose | | 分子名称: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | 著者 | Yang, S, Qin, Z, Zhou, P, Yan, Q, Jiang, Z. | | 登録日 | 2017-03-21 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
4EF5
 
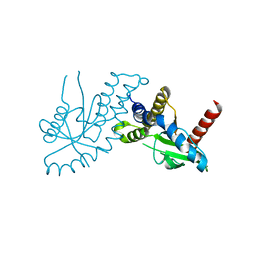 | | Crystal structure of STING CTD | | 分子名称: | Transmembrane protein 173 | | 著者 | Ouyang, S, Ru, H, Shaw, N, Jiang, Y, Niu, F, Zhu, Y, Qiu, W, Li, Y, Liu, Z.-J. | | 登録日 | 2012-03-29 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding
Immunity, 36, 2012
|
|
3UX9
 
 | | Structural insights into a human anti-IFN antibody exerting therapeutic potential for systemic lupus erythematosus | | 分子名称: | Interferon alpha-1/13, ScFv antibody | | 著者 | Ouyang, S, Zhao, L.X, Liang, W, Shaw, N, Liu, Z.-J, Liang, M.-F. | | 登録日 | 2011-12-04 | | 公開日 | 2012-02-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into a human anti-IFN antibody exerting therapeutic potential for systemic lupus erythematosus
J.Mol.Med., 2012
|
|
4EF4
 
 | | Crystal structure of STING CTD complex with c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | 著者 | Ouyang, S, Ru, H, Shaw, N, Jiang, Y, Niu, F, Zhu, Y, Qiu, W, Li, Y, Liu, Z.-J. | | 登録日 | 2012-03-29 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.147 Å) | | 主引用文献 | Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding
Immunity, 36, 2012
|
|
8IPJ
 
 | | Crystal structure of the Legionella effector protein MavL with ADPR-Ub | | 分子名称: | MavL, Ubiquitin, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Ouyang, S, Guan, H. | | 登録日 | 2023-03-14 | | 公開日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Crystal structure of the Legionella effector protein MavL with ADPR-Ub
To Be Published
|
|
8IPW
 
 | | The sturecture of Legionella effector protein MavL with ADPR | | 分子名称: | MavL, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Ouyang, S, Hongxin, G. | | 登録日 | 2023-03-15 | | 公開日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The sturecture of Legionella effector protein MavL with ADPR
To Be Published
|
|
7WQY
 
 | |
7DMS
 
 | |
2GYT
 
 | |
4WY5
 
 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | 分子名称: | Esterase, SULFATE ION | | 著者 | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | 登録日 | 2014-11-15 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
4WY8
 
 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | 分子名称: | esterase | | 著者 | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | 登録日 | 2014-11-16 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
5JLE
 
 | | Crystal structure of SETD2 bound to SAH | | 分子名称: | Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | 著者 | Li, H, Yang, S, Zheng, X. | | 登録日 | 2016-04-27 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase
Genes Dev., 30, 2016
|
|
5JLB
 
 | |
6J9J
 
 | |
5XDM
 
 | |
8H5D
 
 | |
