3NYJ
 
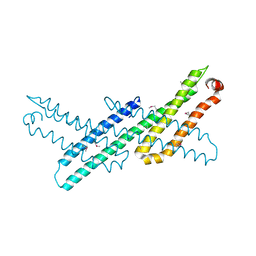 | | Crystal Structure Analysis of APP E2 domain | | Descriptor: | Amyloid beta A4 protein, OSMIUM ION | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
5E3T
 
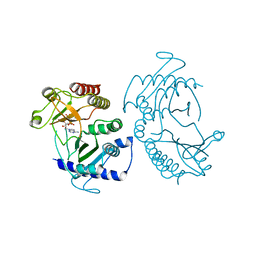 | | Crystal structure of phosphatidylinositol-4-phosphate 5-kinase | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol-4-phosphate 5-kinase, ... | | Authors: | Muftuoglu, Y. | | Deposit date: | 2015-10-04 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanism of substrate specificity of phosphatidylinositol phosphate kinases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5E3S
 
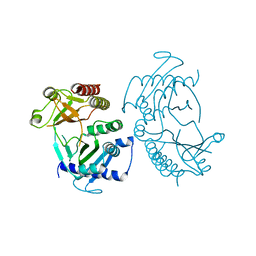 | |
5E3U
 
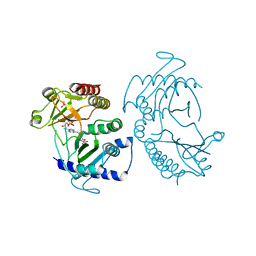 | | Crystal structure of phosphatidylinositol-4-phosphate 5-kinase | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol-4-phosphate 5-kinase, ... | | Authors: | Muftuoglu, Y. | | Deposit date: | 2015-10-04 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanism of substrate specificity of phosphatidylinositol phosphate kinases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GMQ
 
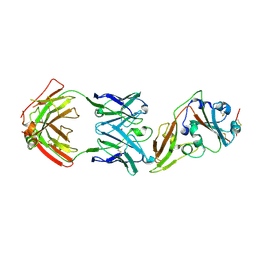 | | Structure of MERS-CoV RBD in complex with a fully human antibody MCA1 | | Descriptor: | 1,2-ETHANEDIOL, MCA1 heavy chain, MCA1 light chain, ... | | Authors: | Chen, C, Wang, J.M, Zou, T.T, Gao, X.P, Cui, S, Jin, Q. | | Deposit date: | 2016-07-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Human Neutralizing Monoclonal Antibody Inhibition of Middle East Respiratory Syndrome Coronavirus Replication in the Common Marmoset.
J. Infect. Dis., 215, 2017
|
|
5IL0
 
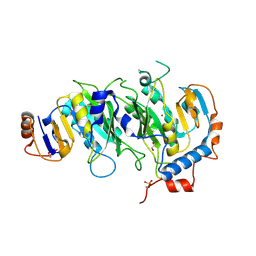 | | Crystal structural of the METTL3-METTL14 complex for N6-adenosine methylation | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, METTL14, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL1
 
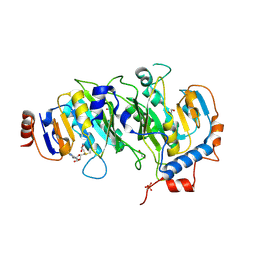 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL2
 
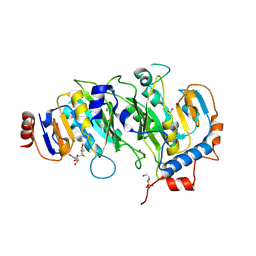 | | Crystal structure of SAH-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
2LSJ
 
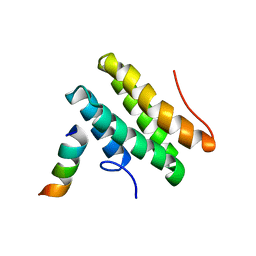 | |
5XIW
 
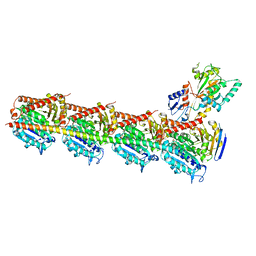 | | Crystal structure of T2R-TTL-Colchicine complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-04-27 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
2LSG
 
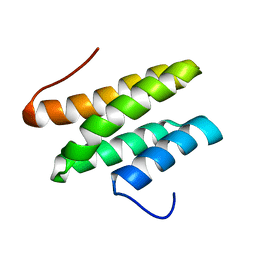 | |
5XP3
 
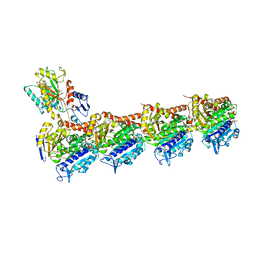 | | Crystal structure of apo T2R-TTL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
5YL2
 
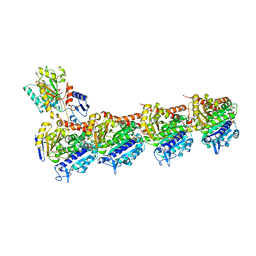 | | Crystal structure of T2R-TTL-Y28 complex | | Descriptor: | (E)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)-3-(4-methoxy-3-oxidanyl-phenyl)prop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yang, J.H, Yang, T, Wen, J.L, Chen, L.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
4CSM
 
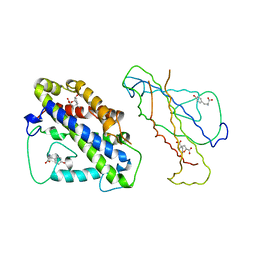 | | YEAST CHORISMATE MUTASE + TYR + ENDOOXABICYCLIC INHIBITOR | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHORISMATE MUTASE, TYROSINE | | Authors: | Straeter, N, Schnappauf, G, Braus, G, Lipscomb, W.N. | | Deposit date: | 1997-07-14 | | Release date: | 1998-01-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures.
Structure, 5, 1997
|
|
4DAY
 
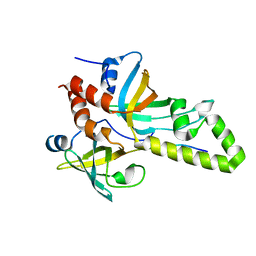 | | Crystal structure of the RMI core complex with MM2 peptide from FANCM | | Descriptor: | Fanconi anemia group M protein, RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | Authors: | Hoadley, K.A, Keck, J.L. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Defining the molecular interface that connects the Fanconi anemia protein FANCM to the Bloom syndrome dissolvasome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2CSM
 
 | |
3CSM
 
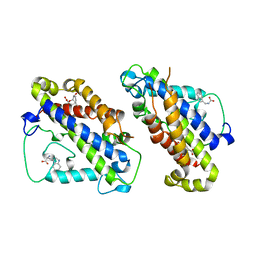 | | STRUCTURE OF YEAST CHORISMATE MUTASE WITH BOUND TRP AND AN ENDOOXABICYCLIC INHIBITOR | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHORISMATE MUTASE, TRYPTOPHAN | | Authors: | Straeter, N, Schnappauf, G, Braus, G, Lipscomb, W.N. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures.
Structure, 5, 1997
|
|
1RDZ
 
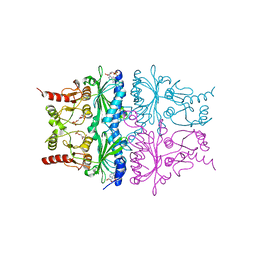 | | T-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
1RDY
 
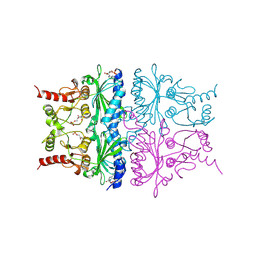 | | T-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
1RDX
 
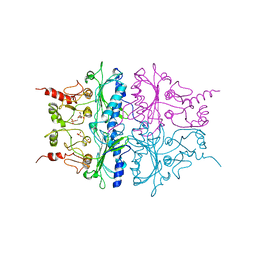 | | R-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
1YLB
 
 | | NMR solution structure of the reduced spinach plastocyanin | | Descriptor: | COPPER (I) ION, Plastocyanin, chloroplast | | Authors: | Musiani, F, Dikiy, A, Semenov, A.Y, Ciurli, S. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the Intermolecular Complex between Plastocyanin and Cytochrome f from Spinach.
J.Biol.Chem., 280, 2005
|
|
4MTE
 
 | |
4MTD
 
 | |
4PIK
 
 | |
4PIT
 
 | |
