3TYU
 
 | |
3TYC
 
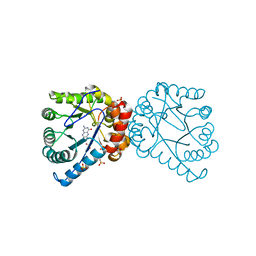 | | Dihydropteroate Synthase in complex with DHP+ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, Dihydropteroate synthase, SULFATE ION | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
3TYD
 
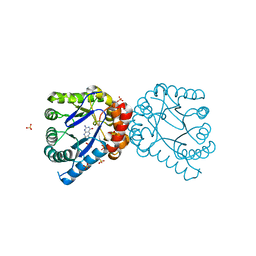 | | Dihydropteroate Synthase in complex with PPi and DHP+ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, Dihydropteroate synthase, PYROPHOSPHATE 2-, ... | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
3UDE
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1B | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine, ACETATE ION | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-28 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
3UD5
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1A | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-27 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
3UDV
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1C | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine, ACETATE ION | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-28 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
5GS4
 
 | | Crystal structure of estrogen receptor alpha in complex with a stabilized peptide antagonist | | Descriptor: | ARG-IAS-ILE-LEU-DNP-ARG-LEU-LEU-GLN, ESTRADIOL, Estrogen receptor, ... | | Authors: | Xie, M, Wang, T, Li, Z.-G. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-30 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Inhibition of ER alpha-Coactivator Interaction by High-Affinity N-Terminus Isoaspartic Acid Tethered Helical Peptides
J. Med. Chem., 60, 2017
|
|
5GTR
 
 | |
2MJV
 
 | |
4QZV
 
 | | Bat-derived coronavirus HKU4 uses MERS-CoV receptor human CD26 for cell entry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Gao, F.G, Wang, Q.H, Qi, J.X, Lu, G.W. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Bat Origins of MERS-CoV Supported by Bat Coronavirus HKU4 Usage of Human Receptor CD26.
Cell Host Microbe, 16, 2014
|
|
8I3U
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 14B1, Light chain of Fab 14B1, ... | | Authors: | Li, Z, Yu, F, Cao, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8I3S
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain od Fab 7B3, Light chain of Fab 7B3, ... | | Authors: | Li, Z, Yu, F, Cao, S, ZHao, H. | | Deposit date: | 2023-01-17 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
7M7X
 
 | |
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7FCQ
 
 | |
7FCP
 
 | |
5WB6
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8P8C
 
 | | HUMAN CD38 ECTODOMAIN BOUND TO COMPOUND 9-ADPR ADDUCT | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[4-[[4-(2-methoxyethoxy)cyclohexyl]amino]-1-methyl-2-oxidanylidene-quinolin-6-yl]pyrazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Rangel, V, Zebisch, M, Doyle, K.J, Burli, R.W. | | Deposit date: | 2023-05-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | A Covalent Binding Mode of a Pyrazole-Based CD38 Inhibitor
Helv.Chim.Acta, 106, 2023
|
|
3P11
 
 | |
3P0Y
 
 | | anti-EGFR/HER3 Fab DL11 in complex with domain III of EGFR extracellular region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2010-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies.
Cancer Cell, 20, 2011
|
|
3P0V
 
 | | anti-EGFR/HER3 Fab DL11 alone | | Descriptor: | CALCIUM ION, Fab DL11 heavy chain, Fab DL11 light chain | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2010-09-29 | | Release date: | 2011-10-19 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies.
Cancer Cell, 20, 2011
|
|
5GYQ
 
 | |
8TN1
 
 | |
8TNB
 
 | |
8TNC
 
 | |
