3OW3
 
 | | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors | | Descriptor: | (2R)-3-(1H-indol-3-yl)-1-{4-[(5S)-5-methyl-5,7-dihydrothieno[3,4-d]pyrimidin-4-yl]piperazin-1-yl}-1-oxopropan-2-amine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Dizon, F, Wu, W, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OW4
 
 | | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors | | Descriptor: | (2R)-3-(1H-indol-3-yl)-1-{4-[(5S)-5-methyl-5,7-dihydrothieno[3,4-d]pyrimidin-4-yl]piperazin-1-yl}-1-oxopropan-2-amine, GSK 3 beta peptide, RAC-alpha serine/threonine-protein kinase | | Authors: | Dizon, F, Wu, W, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5WRV
 
 | | Complex structure of human SRP72/SRP68 | | Descriptor: | ACETATE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Gao, Y, Chen, Z. | | Deposit date: | 2016-12-04 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation
J Mol Cell Biol, 9, 2017
|
|
5WRW
 
 | | Structure of human apo-SRP72 | | Descriptor: | SULFATE ION, Signal recognition particle subunit SRP72 | | Authors: | Gao, Y, Chen, Z. | | Deposit date: | 2016-12-04 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation
J Mol Cell Biol, 9, 2017
|
|
4EEV
 
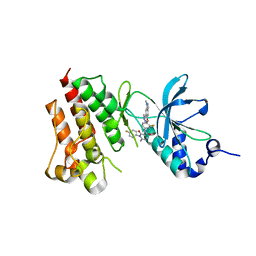 | | Crystal structure of c-Met in complex with LY2801653 | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[1-methyl-6-(1H-pyrazol-4-yl)-1H-indazol-5-yl]oxy}phenyl)-1-(4-fluorophenyl)-6-methyl-2-oxo-1,2-dihydropyridine-3-carboxamide | | Authors: | Wang, Y, Stout, S.L. | | Deposit date: | 2012-03-28 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models.
Invest New Drugs, 31, 2013
|
|
1N2D
 
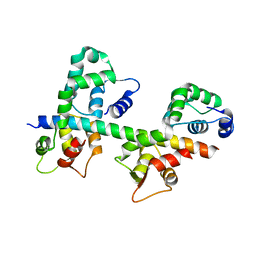 | | Ternary complex of MLC1P bound to IQ2 and IQ3 of Myo2p, a class V myosin | | Descriptor: | IQ2 AND IQ3 MOTIFS FROM MYO2P, A CLASS V MYOSIN, Myosin Light Chain | | Authors: | Terrak, M, Wu, G, Stafford, W.F, Lu, R.C, Dominguez, R. | | Deposit date: | 2002-10-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the light chain-binding domain of myosin V.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | Descriptor: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | Descriptor: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
8FLN
 
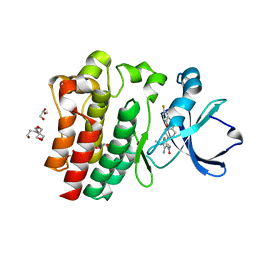 | | Crystal structure of BTK C481S kinase domain in complex with pirtobrutinib | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Cedervall, E.P, Morales, T.H, Allerston, C.K. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.334 Å) | | Cite: | Preclinical characterization of pirtobrutinib, a highly selective, noncovalent (reversible) BTK inhibitor.
Blood, 142, 2023
|
|
8FLL
 
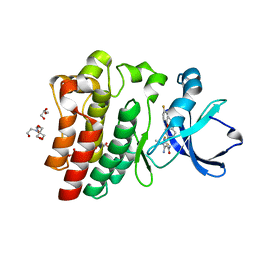 | | Crystal structure of BTK kinase domain in complex with pirtobrutinib | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Cedervall, E.P, Morales, T.H, Allerston, C.K. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Preclinical characterization of pirtobrutinib, a highly selective, noncovalent (reversible) BTK inhibitor.
Blood, 142, 2023
|
|
8HBB
 
 | |
8HB2
 
 | |
8HAZ
 
 | |
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
8HI4
 
 | |
3OQU
 
 | | Crystal structure of native abscisic acid receptor PYL9 with ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL9 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2010-09-04 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
3LD1
 
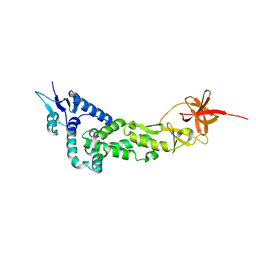 | | Crystal Structure of IBV Nsp2a | | Descriptor: | Replicase polyprotein 1a | | Authors: | Xu, Y, Cong, L, Wei, L, Fu, J, Chen, C, Yang, A, Tang, H, Bartlam, M, Rao, Z. | | Deposit date: | 2010-01-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | IBV nsp2 is an endosome-associated protein and viral pathogenicity factor
To be Published
|
|
2N8A
 
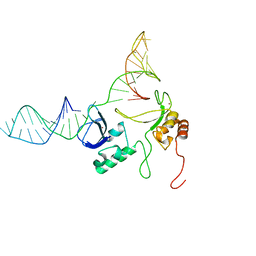 | | 1H, 13C and 15N chemical shift assignments and solution structure for PARP-1 F1F2 domains in complex with a DNA single-strand break | | Descriptor: | DNA (45-MER), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Wu, W. | | Deposit date: | 2015-10-08 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1.
Mol.Cell, 60, 2015
|
|
7EEI
 
 | |
5D0N
 
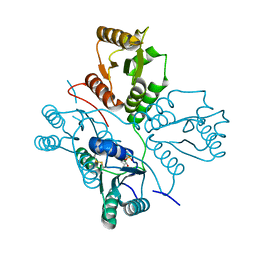 | | Crystal structure of maize PDRP bound with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Pyruvate, ... | | Authors: | Jiang, L, Chen, Z. | | Deposit date: | 2015-08-03 | | Release date: | 2016-02-24 | | Last modified: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein
Plant Physiol., 170, 2016
|
|
5D1F
 
 | | Crystal structure of maize PDRP bound with AMP and Hg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Jiang, L, Chen, Z. | | Deposit date: | 2015-08-04 | | Release date: | 2016-02-24 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein
Plant Physiol., 170, 2016
|
|
8I8C
 
 | |
8I8A
 
 | |
8I8B
 
 | | Outer shell and inner layer structures of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) | | Descriptor: | 38K, AcOrf-109 peptide, Early 49 Daa protein, ... | | Authors: | Jia, X, Gao, Y, Zhang, Q. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Architecture of the baculovirus nucleocapsid revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7XPQ
 
 | |
