7DRI
 
 | | Structure of SspE_CTD_41658 | | Descriptor: | DUF1524 domain | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-28 | | Release date: | 2022-06-29 | | Last modified: | 2023-01-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
5B6D
 
 | |
5B6E
 
 | |
3A08
 
 | | Structure of (PPG)4-OOG-(PPG)4, monoclinic, twinned crystal | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Wu, G, Noguchi, K, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-03-06 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Two crystal modifications of (Pro-Pro-Gly)4-Hyp-Hyp-Gly-(Pro-Pro-Gly)4 reveal the puckering preference of Hyp(X) in the Hyp(X):Hyp(Y) and Hyp(X):Pro(Y) stacking pairs in collagen helices.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A19
 
 | | Structure of (PPG)4-OOG-(PPG)4_H monoclinic, twinned crystal | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Hyakutake, M, Wu, G, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-03-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two crystal modifications of (Pro-Pro-Gly)4-Hyp-Hyp-Gly-(Pro-Pro-Gly)4 reveal the puckering preference of Hyp(X) in the Hyp(X):Hyp(Y) and Hyp(X):Pro(Y) stacking pairs in collagen helices.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6V0L
 
 | | PDGFR-b Promoter Forms a G-Vacancy Quadruplex that Can be Complemented by dGMP: Molecular Structure and Recognition of Guanine Derivatives and Metabolites | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*(3D1)P*AP*GP*GP*GP*AP*GP*GP*GP*CP*GP*GP*CP*GP*GP*GP*AP*CP*A)-3') | | Authors: | Wang, K.B, Dickerhoff, J, Wu, G, Yang, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | SOLUTION NMR | | Cite: | PDGFR-beta Promoter Forms a Vacancy G-Quadruplex that Can Be Filled in by dGMP: Solution Structure and Molecular Recognition of Guanine Metabolites and Drugs.
J.Am.Chem.Soc., 142, 2020
|
|
7CMS
 
 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin | | Descriptor: | (5~{S},6~{R})-5-methyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, PHOSPHATE ION, Tryptophan--tRNA ligase | | Authors: | Lyu, G, Fan, S, Jin, Y, Zou, S, Wu, G, Yang, Z. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the specific interaction between Geobacillus stearothermophilus tryptophanyl-tRNA synthetase and antimicrobial Chuangxinmycin.
J.Biol.Chem., 298, 2022
|
|
6D3Z
 
 | | Protease SFTI complex | | Descriptor: | Plasminogen, Trypsin inhibitor 1 | | Authors: | Law, R.H.P, Wu, G. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Highly Potent and Selective Plasmin Inhibitors Based on the Sunflower Trypsin Inhibitor-1 Scaffold Attenuate Fibrinolysis in Plasma.
J. Med. Chem., 62, 2019
|
|
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
6D40
 
 | |
6D3Y
 
 | |
6D3X
 
 | |
7CKI
 
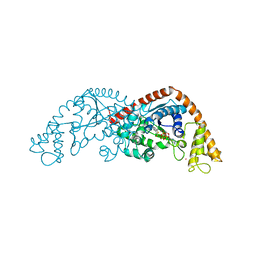 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin and ATP | | Descriptor: | (5~{S},6~{R})-5-methyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lyu, G, Fan, S, Jin, Y, Liu, J, Zou, S, Wu, G, Yang, Z. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin and ATP
To Be Published
|
|
6A4I
 
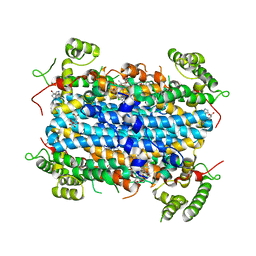 | | Crystal Structure of human TDO inhibitor complex | | Descriptor: | 1-(6-chloro-1H-indazol-4-yl)cyclohexan-1-ol, CITRIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fu, G, Wang, J, Luo, G, Wu, G, Qian, K. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of human TDO inhibitor complex
To Be Published
|
|
4OHR
 
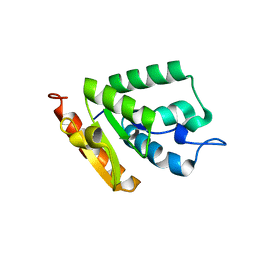 | | Crystal structure of MilB from Streptomyces rimofaciens | | Descriptor: | CMP/hydroxymethyl CMP hydrolase | | Authors: | Zhao, G, Zhang, Y, Liu, G, Wu, G, He, X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry
Nucleic Acids Res., 42, 2014
|
|
7EER
 
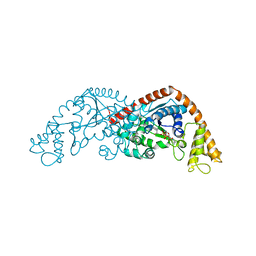 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with 05E6 and ATP | | Descriptor: | 2-(1H-indol-3-yl)ethanol, ADENOSINE-5'-TRIPHOSPHATE, Tryptophan--tRNA ligase | | Authors: | Lv, G, Fan, S, Feng, X, Zhang, Q, Wu, G, Jin, Y, Yang, Z. | | Deposit date: | 2021-03-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with O5E6 and ATP
To Be Published
|
|
4OHB
 
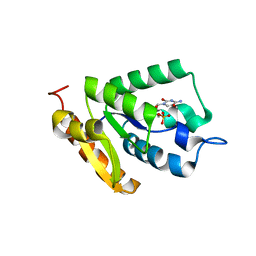 | | Crystal structure of MilB E103A in complex with 5-hydroxymethylcytidine 5'-monophosphate (hmCMP) from Streptomyces rimofaciens | | Descriptor: | 5-(hydroxymethyl)cytidine 5'-(dihydrogen phosphate), CMP/hydroxymethyl CMP hydrolase | | Authors: | Zhao, G, Zhang, Y, Liu, G, Wu, G, He, X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry
Nucleic Acids Res., 42, 2014
|
|
4E57
 
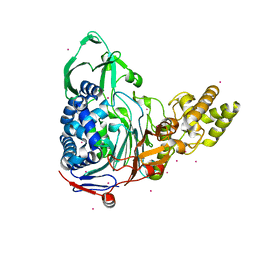 | |
4E55
 
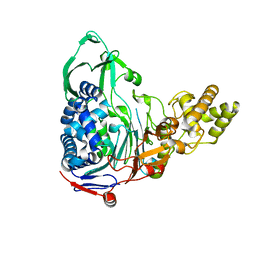 | |
4E56
 
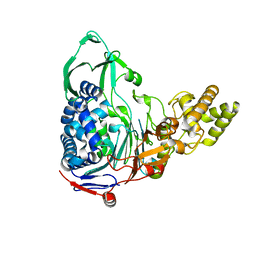 | |
4K6H
 
 | | Crystal structure of CALB mutant L278M from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-15 | | Release date: | 2014-01-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
4K5Q
 
 | |
4K6K
 
 | | Crystal structure of CALB mutant D223G from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-16 | | Release date: | 2014-01-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
4LRV
 
 | | Crystal structure of DndE from Escherichia coli B7A involved in DNA phosphorothioation modification | | Descriptor: | DNA sulfur modification protein DndE | | Authors: | Hu, W, Wang, C.K, Liang, J.D, Zhang, T.L, Yang, M, Hu, Z.P, Wang, Z.J, Lan, W.X, Wu, H.M, Ding, J.P, Wu, G, Deng, Z.X, Cao, C. | | Deposit date: | 2013-07-21 | | Release date: | 2013-08-28 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification
Cell Res., 22, 2012
|
|
1V6Q
 
 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.3 A | | Descriptor: | Collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-12-03 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
