2J37
 
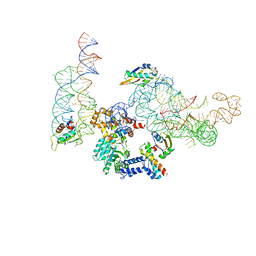 | | MODEL OF MAMMALIAN SRP BOUND TO 80S RNCS | | Descriptor: | 60S RIBOSOMAL PROTEIN L23, RIBOSOMAL PROTEIN L31, RIBOSOMAL PROTEIN L35, ... | | Authors: | Halic, M, Blau, M, Becker, T, Mielke, T, Pool, M.R, Wild, K, Sinning, I, Beckmann, R. | | Deposit date: | 2006-08-18 | | Release date: | 2006-11-08 | | Last modified: | 2019-04-10 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Following the signal sequence from ribosomal tunnel exit to signal recognition particle.
Nature, 444, 2006
|
|
1QZW
 
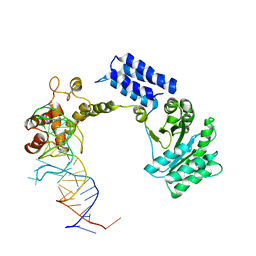 | | Crystal structure of the complete core of archaeal SRP and implications for inter-domain communication | | Descriptor: | 7S RNA, Signal recognition 54 kDa protein | | Authors: | Rosendal, K.R, Wild, K, Montoya, G, Sinning, I. | | Deposit date: | 2003-09-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1QZX
 
 | |
2PX3
 
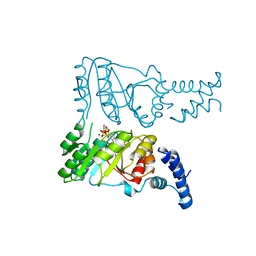 | | Crystal structure of FlhF complexed with GTP/Mg(2+) | | Descriptor: | Flagellar biosynthesis protein flhF, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PX0
 
 | | Crystal structure of FlhF complexed with GMPPNP/Mg(2+) | | Descriptor: | Flagellar biosynthesis protein flhF, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q8K
 
 | | The crystal structure of Ebp1 | | Descriptor: | GLYCEROL, Proliferation-associated protein 2G4, SULFATE ION | | Authors: | Kowalinski, E, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions.
Febs Lett., 581, 2007
|
|
2QY9
 
 | |
3B9Q
 
 | | The crystal structure of cpFtsY from Arabidopsis thaliana | | Descriptor: | Chloroplast SRP receptor homolog, alpha subunit CPFTSY, MALONATE ION | | Authors: | Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2007-11-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of the chloroplast signal recognition particle (SRP) receptor reveals mechanistic details of SRP GTPase activation and a conserved membrane targeting site
Febs Lett., 581, 2007
|
|
3DEO
 
 | |
3DEP
 
 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | Descriptor: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | Authors: | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
3SJB
 
 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJC
 
 | |
3SJA
 
 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJD
 
 | | Crystal structure of S. cerevisiae Get3 with bound ADP-Mg2+ in complex with Get2 cytosolic domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 2, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SYN
 
 | | Crystal structure of FlhF in complex with its activator | | Descriptor: | ALUMINUM FLUORIDE, ATP-binding protein YlxH, Flagellar biosynthesis protein flhF, ... | | Authors: | Bange, G, Kuemmerer, N, Wild, K, Sinning, I. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.063 Å) | | Cite: | Structural basis for the molecular evolution of SRP-GTPase activation by protein.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3UI2
 
 | |
5TBD
 
 | | Crystal Structure of anti-MSP2 Fv fragment (mAb4D11) in complex with 3D7-MSP2 215-222 | | Descriptor: | Fv fragment (mAb4D1) heavy chain, Merozoite surface protein 2 | | Authors: | Seow, J, Morales, R.A.V, MacRaild, C.A, Bankala, K, Drinkwater, N, Dingjan, T, Jaipuria, G, Wilde, K, Anders, R.F, Atreya, H.S, Christ, D, McGowan, S, Norton, R.S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Characterisation of a Key Epitope in the Conserved C-Terminal Domain of the Malaria Vaccine Candidate MSP2.
J. Mol. Biol., 429, 2017
|
|
3KL4
 
 | | Recognition of a signal peptide by the signal recognition particle | | Descriptor: | Signal peptide of yeast dipeptidyl aminopeptidase B, Signal recognition 54 kDa protein | | Authors: | Janda, C.Y, Nagai, K, Li, J, Oubridge, C. | | Deposit date: | 2009-11-06 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Recognition of a signal peptide by the signal recognition particle.
Nature, 465, 2010
|
|
1E2K
 
 | | Kinetics and crystal structure of the wild-type and the engineered Y101F mutant of Herpes simplex virus type 1 thymidine kinase interacting with (North)-methanocarba-thymidine | | Descriptor: | 1-[4-HYDROXY-5-(HYDROXYMETHYL)BICYCLO[3.1.0]HEX-2-YL]-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-08-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetics and Crystal Structure of the Wild-Type and the Engineered Y101F Mutant of Herpes Simplex Virus Type 1 Thymidine Kinase Interacting with (North)-Methanocarba-Thymidine
Biochemistry, 39, 2000
|
|
1E2L
 
 | | Kinetics and crystal structure of the wild-type and the engineered Y101F mutant of Herpes simplex virus type 1 thymidine kinase interacting with (North)-methanocarba-thymidine | | Descriptor: | 1-[4-HYDROXY-5-(HYDROXYMETHYL)BICYCLO[3.1.0]HEX-2-YL]-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetics and Crystal Structure of the Wild-Type and the Engineered Y101F Mutant of Herpes Simplex Virus Type 1 Thymidine Kinase Interacting with (North)-Methanocarba-Thymidine
Biochemistry, 39, 2000
|
|
2N88
 
 | | Chromodomain 3 (CD3) of cpSRP43 | | Descriptor: | Signal recognition particle 43 kDa protein, chloroplastic | | Authors: | Hennig, J, Sattler, M. | | Deposit date: | 2015-10-06 | | Release date: | 2015-12-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
1OF1
 
 | | KINETICS AND CRYSTAL STRUCTURE OF THE HERPES SIMPLEX VIRUS TYPE 1 THYMIDINE KINASE INTERACTING WITH (SOUTH)-METHANOCARBA-THYMIDINE | | Descriptor: | (SOUTH)-METHANOCARBA-THYMIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Claus, M.T, Schelling, P, Folkers, G, Marquez, V.E, Scapozza, L, Schulz, G.E. | | Deposit date: | 2003-04-03 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and Structural Characterization of (South)-Methanocarbathymidine that Specifically Inhibits Growth of Herpes Simplex Virus Type 1 Thymidine Kinase-Transduced Osteosarcoma Cells
J.Biol.Chem., 279, 2004
|
|
3BS6
 
 | |
1E2N
 
 | | HPT + HMTT | | Descriptor: | 6-{[4-(HYDROXYMETHYL)-5-METHYL-2,6-DIOXOHEXAHYDROPYRIMIDIN-5-YL]METHYL}-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2001-03-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Effect of Substrate Binding on the Conformation and Structural Stability of Herpes Simplex Virus Type 1 Thymidine Kinase
Protein Sci., 10, 2001
|
|
1E2H
 
 | | The nucleoside binding site of Herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography | | Descriptor: | SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleoside Binding Site of Herpes Simplex Type 1 Thymidine Kinase Analyzed by X-Ray Crystallography
Proteins: Struct.,Funct., Genet., 41, 2000
|
|
