3DWP
 
 | | Crystal structure of the B-subunit of the AB5 toxin from E. Coli with Neu5Gc | | Descriptor: | N-glycolyl-alpha-neuraminic acid, PENTAETHYLENE GLYCOL, Subtilase cytotoxin, ... | | Authors: | Byres, E, Paton, A.W, Paton, J.C, Lofling, J.C, Smith, D.F, Wilce, M.C.J, Talbot, U.M, Chong, D.C, Yu, H, Huang, S, Chen, X, Varki, N.M, Varki, A, Rossjohn, J, Beddoe, T. | | Deposit date: | 2008-07-22 | | Release date: | 2008-11-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin
Nature, 456, 2008
|
|
1AV4
 
 | |
1AVK
 
 | | CRYSTAL STRUCTURES OF THE COPPER-CONTAINING AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS IN THE HOLO-AND APO-FORMS: IMPLICATIONS FOR THE BIOGENESIS OF TOPA QUINONE | | Descriptor: | AMINE OXIDASE | | Authors: | Wilce, M.C.J, Guss, J.M, Freeman, H.C, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 1997-09-16 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the copper-containing amine oxidase from Arthrobacter globiformis in the holo and apo forms: implications for the biogenesis of topaquinone.
Biochemistry, 36, 1997
|
|
1AVL
 
 | |
1FS0
 
 | |
6ORU
 
 | |
1JAW
 
 | | AMINOPEPTIDASE P FROM E. COLI LOW PH FORM | | Descriptor: | ACETATE ION, AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Wilce, M.C.J, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1KSI
 
 | | CRYSTAL STRUCTURE OF A EUKARYOTIC (PEA SEEDLING) COPPER-CONTAINING AMINE OXIDASE AT 2.2A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Wilce, M.C.J, Kumar, V, Freeman, H.C, Guss, J.M. | | Deposit date: | 1996-07-20 | | Release date: | 1997-12-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2 A resolution.
Structure, 4, 1996
|
|
5ITH
 
 | |
5U06
 
 | | Grb7-SH2 with bicyclic peptide inhibitor containing a pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, POTASSIUM ION, bicyclic peptide inhibitor: LYS-PHE-GLU-GLY-CMF-ASP-ASN-GLU-CST | | Authors: | Watson, G.M, Wilce, J.A. | | Deposit date: | 2016-11-22 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
4TZG
 
 | | Crystal structure of eCGP123, an extremely thermostable green fluorescent protein | | Descriptor: | Fluorescent Protein | | Authors: | Close, D.W, Don Paul, C, Traore, D.A.K, Wilce, M.C.J, Prescott, M, Bradbury, A.R.M. | | Deposit date: | 2014-07-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermal green protein, an extremely stable, nonaggregating fluorescent protein created by structure-guided surface engineering.
Proteins, 83, 2015
|
|
5EEQ
 
 | | Grb7 SH2 with the G7-B1 bicyclic peptide inhibitor | | Descriptor: | Bicyclic Peptide Inhibitor, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Ambaye, N.D, Watson, G.M, Wilce, M.C.J, Wilce, G.M. | | Deposit date: | 2015-10-23 | | Release date: | 2016-06-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
5D0J
 
 | | Grb7 SH2 with inhibitor peptide | | Descriptor: | G7-TEdFP peptide, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Gunzburg, M.J, Watson, G.M, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2015-08-03 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
5EEL
 
 | | Grb7 SH2 with bicyclic peptide inhibitor | | Descriptor: | Bicyclic Peptide Inhibitor, FORMIC ACID, Growth factor receptor-bound protein 7, ... | | Authors: | Watson, G.M, Gunzburg, M.J, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2015-10-23 | | Release date: | 2016-06-15 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
4WOP
 
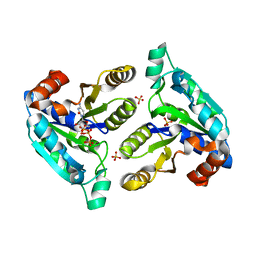 | | Nucleotide Triphosphate Promiscuity in Mycobacterium tuberculosis Dethiobiotin Synthetase | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-TRIPHOSPHATE, SULFATE ION | | Authors: | Salaemae, W, Yap, M.Y, Wegener, K.L, Booker, G.W, Wilce, M.C.J, Polyak, S.W. | | Deposit date: | 2014-10-16 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Nucleotide triphosphate promiscuity in Mycobacterium tuberculosis dethiobiotin synthetase.
Tuberculosis (Edinb), 95, 2015
|
|
6NDL
 
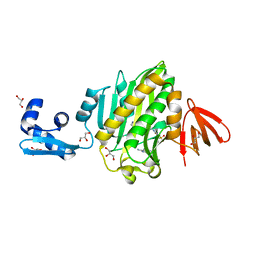 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with a sulfonamide inhibitor | | Descriptor: | 1-[4-(6-aminopurin-9-yl)butylsulfamoyl]-3-[4-[(4~{S})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butyl]urea, Biotin Protein Ligase, GLYCEROL | | Authors: | Marshall, A.C, Polyak, S.W, Bruning, J.B, Lee, K. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfonamide-Based Inhibitors of Biotin Protein Ligase as New Antibiotic Leads.
Acs Chem.Biol., 14, 2019
|
|
1BE7
 
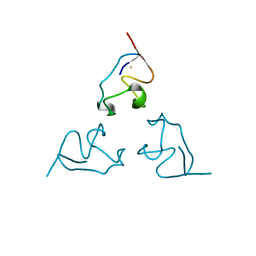 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN C42S MUTANT | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Maher, M, Guss, J.M, Wilce, M, Wedd, A.G. | | Deposit date: | 1998-05-20 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Rubredoxin from Clostridium Pasteurianum: Mutation of the Iron Cysteinyl Ligands to Serine. Crystal and Molecular Structures of the Oxidised and Dithionite-Treated Forms of the Cys42Ser Mutant
J.Am.Chem.Soc., 120, 1998
|
|
3OF6
 
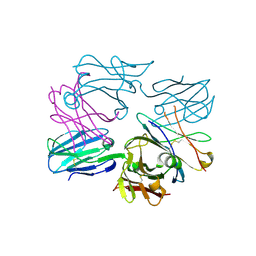 | | Human pre-T cell receptor crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre T-cell antigen receptor alpha, T cell receptor beta chain | | Authors: | Pang, S.S. | | Deposit date: | 2010-08-13 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for autonomous dimerization of the pre-T-cell antigen receptor
Nature, 467, 2010
|
|
2ZNM
 
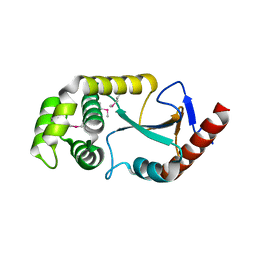 | | Oxidoreductase NmDsbA3 from Neisseria meningitidis | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Vivian, J.P, Scoullar, J, Robertson, A.L, Bottomley, S.P, Horne, J, Chin, Y, Velkov, T, Wielens, J, Thompson, P.E, Piek, S, Byres, E, Beddoe, T, Wilce, M.C.J, Kahler, C, Rossjohn, J, Scanlon, M.J. | | Deposit date: | 2008-04-30 | | Release date: | 2008-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Oxidoreductase NmDsbA3 from Neisseria meningitidis
J.Biol.Chem., 283, 2008
|
|
4PPK
 
 | | Crystal structure of eCGP123 T69V variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
4PPJ
 
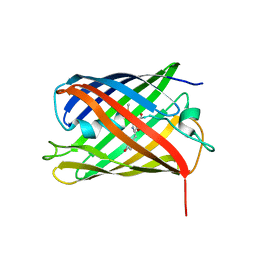 | | Crystal structure of Phanta, a weakly fluorescent photochromic GFP-like protein. ON state | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
4PPL
 
 | | Crystal structure of eCGP123 H193Q variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
1F4K
 
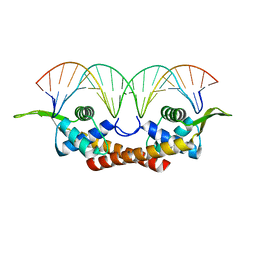 | | CRYSTAL STRUCTURE OF THE REPLICATION TERMINATOR PROTEIN/B-SITE DNA COMPLEX | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*AP*AP*CP*AP*TP*AP*AP*TP*GP*TP*TP*CP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*GP*AP*AP*CP*AP*TP*TP*AP*TP*GP*TP*TP*CP*AP*TP*AP*G)-3', REPLICATION TERMINATION PROTEIN | | Authors: | Wilce, J.A, Vivian, J.P, Hastings, A.F, Otting, G, Folmer, R.H.A, Duggin, I.G, Wake, R.G, Wilce, M.C.J. | | Deposit date: | 2000-06-08 | | Release date: | 2001-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest
Nat.Struct.Biol., 8, 2001
|
|
1B13
 
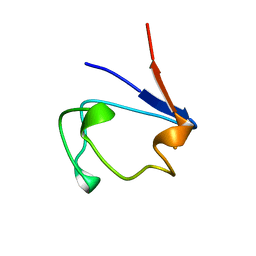 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-26 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2O
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10VG43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-30 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
