2WOU
 
 | | ALK5 IN COMPLEX WITH 4-((4-((2,6-dimethyl-3-pyridyl)oxy)-2-pyridyl) amino)benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(2,6-DIMETHYLPYRIDIN-3-YL)OXY]PYRIDIN-2-YL}AMINO)BENZENESULFONAMIDE, TGF-BETA RECEPTOR TYPE-1 | | Authors: | Debreczeni, J.E, Norman, R.A, Goldberg, F.W, Ward, R.A, Finlay, R, Powell, S.J, Roberts, N.J, Dishington, A.P, Gingell, H.J, Wickson, K.F, Roberts, A.L. | | Deposit date: | 2009-07-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid Generation of a High Quality Lead for Transforming Growth Factor-Beta (Tgf-Beta) Type I Receptor (Alk5).
J.Med.Chem., 52, 2009
|
|
2WOT
 
 | | ALK5 IN COMPLEX WITH 4-((5,6-dimethyl-2-(2-pyridyl)-3-pyridyl)oxy)-N-(3,4,5-trimethoxyphenyl)pyridin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5,6-DIMETHYL-2,2'-BIPYRIDIN-3-YL)OXY]-N-(3,4,5-TRIMETHOXYPHENYL)PYRIDIN-2-AMINE, TGF-BETA RECEPTOR TYPE-1 | | Authors: | Norman, R.A, Debreczeni, J.E, Goldberg, F.W, Ward, R.A, Finlay, R, Powell, S.J, Roberts, N.J, Dishington, A.P, Gingell, H.J, Wickson, K.F, Roberts, A.L. | | Deposit date: | 2009-07-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Rapid Generation of a High Quality Lead for Transforming Growth Factor-Beta (Tgf-Beta) Type I Receptor (Alk5).
J.Med.Chem., 52, 2009
|
|
4D4L
 
 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 5-(4-chlorophenyl)-7-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
4D4K
 
 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, 7-(4-methoxyphenyl)-5-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
4D2S
 
 | | Human TTK in complex with a Dyrk1B inhibitor | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, N-{2-methoxy-4-[(1-methylpiperidin-4-yl)oxy]phenyl}-4-(1H-pyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine | | Authors: | Debreczeni, J.E, Kettle, J.G, Ballard, P, Bardelle, C, Butterworth, S, Colclough, N, Critchlow, S.E, Fairley, G, Fillery, S, Graham, M.A, Goodwin, L, Guichard, S, Hudson, K, Mahmood, A, Vincent, J, Ward, R.A, Whittaker, D. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Optimization of a Novel Series of Dyrk1B Kinase Inhibitors to Explore a Mek Resistance Hypothesis.
J.Med.Chem., 58, 2015
|
|
4D2R
 
 | | Human IGF in complex with a Dyrk1B inhibitor | | Descriptor: | CHLORIDE ION, INSULIN-LIKE GROWTH FACTOR 1 RECEPTOR, N-{2-methoxy-4-[(1-methylpiperidin-4-yl)oxy]phenyl}-4-(1H-pyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine | | Authors: | Debreczeni, J.E, Kettle, J.G, Ballard, P, Bardelle, C, Butterworth, S, Colclough, N, Critchlow, S.E, Fairley, G, Fillery, S, Graham, M.A, Goodwin, L, Guichard, S, Hudson, K, Mahmood, A, Vincent, J, Ward, R.A, Whittaker, D. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of a Novel Series of Dyrk1B Kinase Inhibitors to Explore a Mek Resistance Hypothesis.
J.Med.Chem., 58, 2015
|
|
4D4M
 
 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, 7-(4-bromophenyl)-5-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
4D4J
 
 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 5-(4-bromophenyl)-7-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
9BIV
 
 | |
8UK7
 
 | |
4WHV
 
 | | E3 ubiquitin-protein ligase RNF8 in complex with Ubiquitin-conjugating enzyme E2 N and Polyubiquitin-B | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-09-23 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (8.3 Å) | | Cite: | RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J.Biol.Chem., 291, 2016
|
|
8D34
 
 | |
1IXB
 
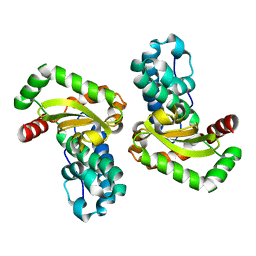 | | CRYSTAL STRUCTURE OF THE E. COLI MANGANESE(II) SUPEROXIDE DISMUTASE MUTANT Y174F AT 0.90 ANGSTROMS RESOLUTION. | | Descriptor: | MANGANESE ION, 1 HYDROXYL COORDINATED, SUPEROXIDE DISMUTASE | | Authors: | Anderson, B.F, Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2002-06-18 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures at 0.90 A resolution of the oxidised and reduced forms of the Y174F mutant of the manganese superoxide dismutase from Escherichia coli
To be Published
|
|
1IX9
 
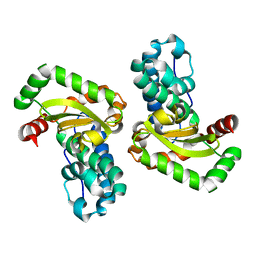 | | Crystal Structure of the E. coli Manganase(III) superoxide dismutase mutant Y174F at 0.90 angstroms resolution. | | Descriptor: | MANGANESE (II) ION, Superoxide Dismutase | | Authors: | Anderson, B.F, Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2002-06-17 | | Release date: | 2002-12-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures at 0.90 A resolution of the oxidised and reduced forms of the Y174F mutant of the manganese superoxide dismutase from Escherichia coli
To be Published
|
|
2ADO
 
 | | Crystal Structure Of The Brct Repeat Region From The Mediator of DNA damage checkpoint protein 1, MDC1 | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Lee, M.S, Edwards, R.A, Thede, G.L, Glover, J.N. | | Deposit date: | 2005-07-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the BRCT Repeat Domain of MDC1 and Its Specificity for the Free COOH-terminal End of the {gamma}-H2AX Histone Tail.
J.Biol.Chem., 280, 2005
|
|
2R1Z
 
 | |
3K16
 
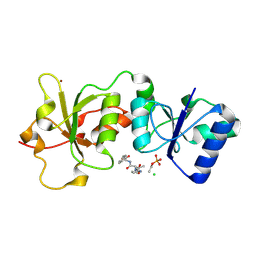 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with a free carboxy C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3K0H
 
 | | The crystal structure of BRCA1 BRCT in complex with a minimal recognition tetrapeptide with an amidated C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3K15
 
 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with an amidated C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
7RGT
 
 | | The crystal structure of RocC, containing FinO domain, 1-126 | | Descriptor: | Repressor of competence, RNA Chaperone, SULFATE ION | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
7RGS
 
 | | The crystal structure of RocC, containing FinO domain, 24-126 | | Descriptor: | Repressor of competence, RNA Chaperone | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
7RGU
 
 | |
3K05
 
 | |
3K0K
 
 | | Crystal Structure of BRCA1 BRCT in complex with a minimal recognition tetrapeptide with a free carboxy C-terminus. | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3OMY
 
 | | Crystal structure of the pED208 TraM N-terminal domain | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein traM | | Authors: | Wong, J.J.W, Lu, J, Edwards, R.A, Frost, L.S, Mark Glover, J.N. | | Deposit date: | 2010-08-27 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of cooperative DNA recognition by the plasmid conjugation factor, TraM.
Nucleic Acids Res., 39, 2011
|
|
