7VHM
 
 | |
7Y8R
 
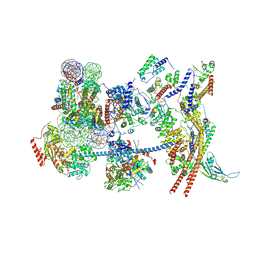 | | The nucleosome-bound human PBAF complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2, ... | | Authors: | Wang, L, Yu, J, Yu, Z, Wang, Q, He, S, Xu, Y. | | Deposit date: | 2022-06-24 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of nucleosome-bound human PBAF complex.
Nat Commun, 13, 2022
|
|
6AD3
 
 | |
6BFN
 
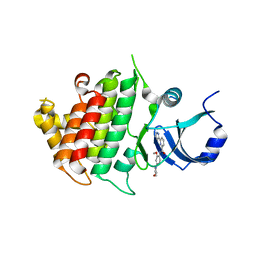 | | Crystal structure of human IRAK1 | | Descriptor: | Interleukin-1 receptor-associated kinase 1, N-[2-methoxy-4-(morpholin-4-yl)phenyl]-6-(1H-pyrazol-5-yl)pyridine-2-carboxamide | | Authors: | Wang, L, Qiao, Q, Wu, H. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of human IRAK1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7Y0O
 
 | |
7Y0N
 
 | | SARS-CoV-2 WT Spike in complex with R15 Fab and P14 Nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R15-F7, ... | | Authors: | Wang, L. | | Deposit date: | 2022-06-05 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 WT Spike in complex with R15 Fab and P14 Nanobody
To Be Published
|
|
6K5E
 
 | | Crystal structure of BioH from Klebsiella pneumonia | | Descriptor: | Pimeloyl-[acyl-carrier protein] methyl ester esterase | | Authors: | Wang, L, Chen, Y. | | Deposit date: | 2019-05-28 | | Release date: | 2019-07-17 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (2.257 Å) | | Cite: | Structural insight into the carboxylesterase BioH from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
6KG7
 
 | | Cryo-EM Structure of the Mammalian Tactile Channel Piezo2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Piezo-type mechanosensitive ion channel component 2 | | Authors: | Wang, L, Zhou, H, Zhang, M, Liu, W, Deng, T, Zhao, Q, Li, Y, Lei, J, Li, X, Xiao, B. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanogating of the mammalian tactile channel PIEZO2.
Nature, 573, 2019
|
|
7YR4
 
 | |
6L3E
 
 | | Crystal structure of Salmonella enterica sugar-binding protein MalE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Salmonella enterica sugar-binding protein MalE
To Be Published
|
|
6L0Z
 
 | | The crystal structure of Salmonella enterica sugar-binding protein MalE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-anhydro-D-glucitol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Salmonella enterica sugar-binding protein MalE
To Be Published
|
|
6L19
 
 | | The crystal structure of competence or damage-inducible protein from Enterobacter asburiae | | Descriptor: | CHLORIDE ION, GLYCEROL, PncC family amidohydrolase, ... | | Authors: | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | Deposit date: | 2019-09-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The crystal structure of Competence or damage-inducible protein from Enterobacter asburiae
To Be Published
|
|
6MIX
 
 | | Human TRPM2 ion channel in apo state | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and gating mechanism of human TRPM2.
Science, 362, 2018
|
|
6MJ2
 
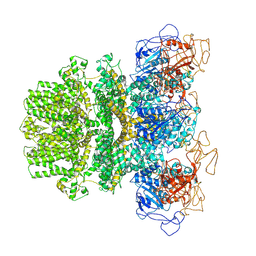 | | Human TRPM2 ion channel in a calcium- and ADPR-bound state | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-02 | | Method: | ELECTRON MICROSCOPY (6.36 Å) | | Cite: | Structures and gating mechanism of human TRPM2.
Science, 362, 2018
|
|
6MIZ
 
 | |
7ZYU
 
 | |
4V9C
 
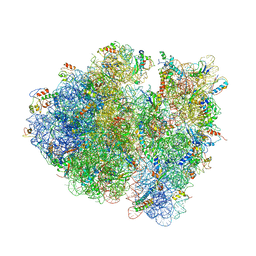 | | Allosteric control of the ribosome by small-molecule antibiotics | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Cate, J.H.D, Pulk, A, Blanchard, S.C, Wang, L, Feldman, M.B, Wasserman, M.R, Altman, R. | | Deposit date: | 2012-07-25 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Allosteric control of the ribosome by small-molecule antibiotics.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4U7P
 
 | | Crystal structure of DNMT3A-DNMT3L complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, L, Guo, X, Li, J, Xiao, J, Yin, X, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.821 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
4X67
 
 | | Crystal structure of elongating yeast RNA polymerase II stalled at oxidative Cyclopurine DNA lesions. | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2014-12-07 | | Release date: | 2015-02-04 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X6A
 
 | | Crystal structure of yeast RNA polymerase II encountering oxidative Cyclopurine DNA lesions | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2014-12-07 | | Release date: | 2015-02-04 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6ND0
 
 | |
8GQU
 
 | | AK-42 inhibitor binding human ClC-2 TMD | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nat Commun, 14, 2023
|
|
7FAP
 
 | | Structure of VAR2CSA-CSA 3D7 | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-3)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Wang, Z. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
7FAS
 
 | | VAR2CSA 3D7 ectodomain core region | | Descriptor: | Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Zhaoning, W. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
3O6Y
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J.K, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-29 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
