5F1B
 
 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GP1, GP2, ... | | 著者 | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | 登録日 | 2015-11-30 | | 公開日 | 2016-01-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
5F18
 
 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | 分子名称: | Niemann-Pick C1 protein | | 著者 | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | 登録日 | 2015-11-30 | | 公開日 | 2016-01-20 | | 最終更新日 | 2016-01-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
4NND
 
 | |
4O4D
 
 | | Crystal Structure of an Inositol hexakisphosphate kinase EhIP6KA in complexed with ATP and Ins(1,4,5)P3 | | 分子名称: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | 著者 | Wang, H, Shears, S.B. | | 登録日 | 2013-12-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family.
Nat Commun, 5, 2014
|
|
4NZM
 
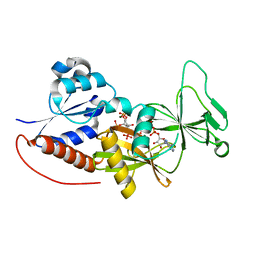 | | Crystal structure of the catalytic domain of PPIP5K2 in complex with AMPPNP and 5-PA-InsP5 | | 分子名称: | (2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]oxy}ethyl)phosphonic acid, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, MAGNESIUM ION, ... | | 著者 | Wang, H, Shears, S.B. | | 登録日 | 2013-12-12 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Synthetic Inositol Phosphate Analogs Reveal that PPIP5K2 Has a Surface-Mounted Substrate Capture Site that Is a Target for Drug Discovery.
Chem.Biol., 21, 2014
|
|
4NZO
 
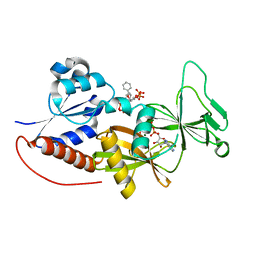 | | Crystal structure of the catalytic domain of PPIP5K2 in complex with AMPPNP and 2,5-DI-O-BN-INSP4 | | 分子名称: | (1R,2R,3r,4S,5S,6s)-3,6-bis(benzyloxy)cyclohexane-1,2,4,5-tetrayl tetrakis[dihydrogen (phosphate)], Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, MAGNESIUM ION, ... | | 著者 | Wang, H, Shears, S.B. | | 登録日 | 2013-12-12 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Synthetic Inositol Phosphate Analogs Reveal that PPIP5K2 Has a Surface-Mounted Substrate Capture Site that Is a Target for Drug Discovery.
Chem.Biol., 21, 2014
|
|
4O4F
 
 | |
4O4C
 
 | |
4O4B
 
 | |
4NZN
 
 | | Crystal structure of the catalytic domain of PPIP5K2 in complex with AMPPNP and 2-O-BN-5-PA-INSP4 | | 分子名称: | (2-{[(1s,2R,3R,4r,5S,6S)-4-(benzyloxy)-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl]oxy}-2-oxoethyl)phosphonic acid, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, MAGNESIUM ION, ... | | 著者 | Wang, H, Shears, S.B. | | 登録日 | 2013-12-12 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Synthetic Inositol Phosphate Analogs Reveal that PPIP5K2 Has a Surface-Mounted Substrate Capture Site that Is a Target for Drug Discovery.
Chem.Biol., 21, 2014
|
|
4O4E
 
 | | Crystal Structure of an Inositol hexakisphosphate kinase EhIP6KA in complexed with ATP and Ins(1,3,4,5,6)P5 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Inositol hexakisphosphate kinase, MAGNESIUM ION, ... | | 著者 | Wang, H, Shears, S.B. | | 登録日 | 2013-12-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family.
Nat Commun, 5, 2014
|
|
5J0F
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P4/5 | | 分子名称: | GLYCEROL, Superoxide dismutase [Cu-Zn],OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | 著者 | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | 登録日 | 2016-03-28 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
5J0G
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P7/8 | | 分子名称: | OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | 著者 | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | 登録日 | 2016-03-28 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
5J07
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P1/2 | | 分子名称: | Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn] | | 著者 | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | 登録日 | 2016-03-27 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
5J0C
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P2/3 | | 分子名称: | Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | 著者 | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | 登録日 | 2016-03-28 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
2NZ0
 
 | | Crystal structure of potassium channel Kv4.3 in complex with its regulatory subunit KChIP1 | | 分子名称: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | 著者 | Wang, H, Yan, Y, Shen, Y, Chen, L, Wang, K. | | 登録日 | 2006-11-22 | | 公開日 | 2006-12-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits.
Nat.Neurosci., 10, 2007
|
|
8HE9
 
 | | Crystal structure of CTSB in complex with K777 | | 分子名称: | Cathepsin B, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Wang, H, Li, D, Sun, L, Yang, H. | | 登録日 | 2022-11-07 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of CTSB in complex with K777
To Be Published
|
|
8HEN
 
 | | Crystal structure of CTSB in complex with 212-148 | | 分子名称: | 2-[4-[[(2~{S})-1-oxidanylidene-3-phenyl-1-[[(3~{S})-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]amino]propan-2-yl]carbamoyl]piperazin-1-yl]ethyl 4-carbamimidamidobenzoate, Cathepsin B, DIMETHYL SULFOXIDE, ... | | 著者 | Wang, H, Li, D, Sun, L, Yang, H. | | 登録日 | 2022-11-08 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of CTSB in complex with 212-148
To Be Published
|
|
8HFV
 
 | | Crystal structure of CTSL in complex with K777 | | 分子名称: | CACODYLATE ION, Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide, Procathepsin L, ... | | 著者 | Wang, H, Shao, M, Sun, L, Yang, H. | | 登録日 | 2022-11-12 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of CTSL in complex with K777
To Be Published
|
|
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | 著者 | Wang, H, Liu, X, Sun, L, Yang, H. | | 登録日 | 2022-11-03 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of TMPRSS2 in complex with 212-148
To Be Published
|
|
8HET
 
 | |
8HEI
 
 | |
8IQL
 
 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | 分子名称: | Apoptosis regulator Bcl-2, Bcl-2-modifying factor | | 著者 | Wang, H, Guo, M, Wei, H, Chen, Y. | | 登録日 | 2023-03-16 | | 公開日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.9577 Å) | | 主引用文献 | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
8IQK
 
 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | 分子名称: | Bcl-2-like protein 1, Bcl-2-modifying factor | | 著者 | Wang, H, Guo, M, Wei, H, Chen, Y. | | 登録日 | 2023-03-16 | | 公開日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.879 Å) | | 主引用文献 | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
8IQM
 
 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | 分子名称: | Bcl2 modifying factor, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Wang, H, Guo, M, Wei, H, Chen, Y. | | 登録日 | 2023-03-16 | | 公開日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.967 Å) | | 主引用文献 | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
