7ROI
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 12 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROC
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 7 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7RO4
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 3 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROD
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 8 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROB
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 6 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7RO5
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 4 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROE
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 9 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7E9W
 
 | | The Crystal Structure of D-psicose-3-epimerase from Biortus. | | Descriptor: | D-psicose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Wang, F, Xu, C, Qi, J, Zhang, M, Tian, F, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of D-psicose-3-epimerase from Biortus.
To Be Published
|
|
5GWY
 
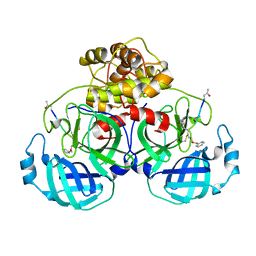 | | Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, main protease | | Authors: | Wang, F, Chen, C, Tan, W, Yang, K, Yang, H. | | Deposit date: | 2016-09-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design.
Sci Rep, 6, 2016
|
|
7TGG
 
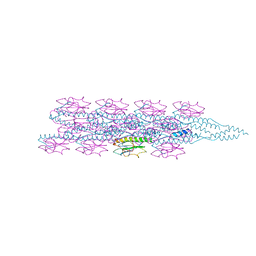 | | Cryo-EM structure of PilA-N and PilA-C from Geobacter sulfurreducens | | Descriptor: | Geopilin domain 1 protein, Geopilin domain 2 protein | | Authors: | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Bond, D.R, Hochbaum, A.I, Egelman, E.H. | | Deposit date: | 2022-01-07 | | Release date: | 2022-02-23 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
7TFS
 
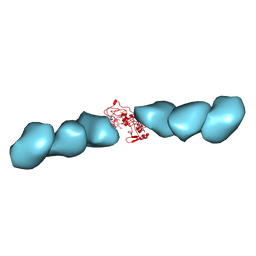 | | Cryo-EM of the OmcE nanowires from Geobacter sulfurreducens | | Descriptor: | Cytochrome c, HEME C | | Authors: | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Hochbaum, A.I, Bond, D.R, Egelman, E.H. | | Deposit date: | 2022-01-07 | | Release date: | 2022-05-04 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
7TXJ
 
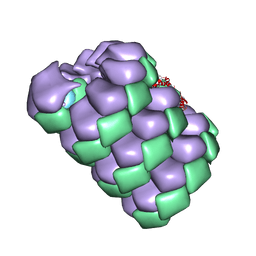 | |
7UEG
 
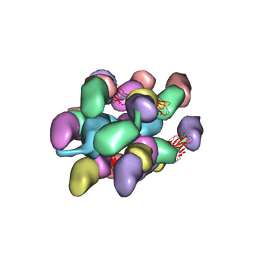 | |
7UIT
 
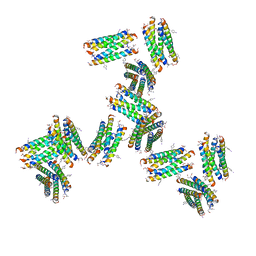 | |
7UII
 
 | |
7UQJ
 
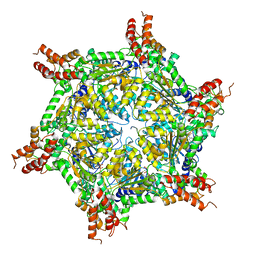 | | Cryo-EM structure of the S. cerevisiae chromatin remodeler Yta7 hexamer bound to ATPgS and histone H3 tail in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase histone chaperone YTA7, Histone H3, ... | | Authors: | Wang, F, Feng, X, Li, H. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-01 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Saccharomyces cerevisiae Yta7 ATPase hexamer contains a unique bromodomain tier that functions in nucleosome disassembly.
J.Biol.Chem., 299, 2022
|
|
7UQK
 
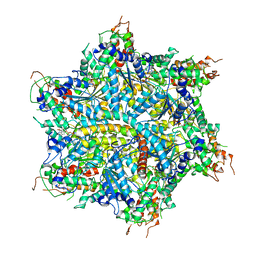 | |
7UQI
 
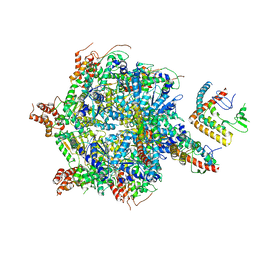 | |
5JR3
 
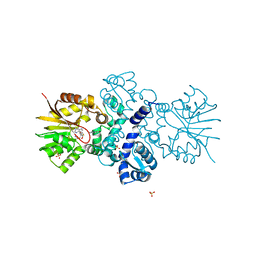 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Johnson, B.R, Huber, T.D, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-05-05 | | Release date: | 2016-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone (to be published)
To Be Published
|
|
2QEG
 
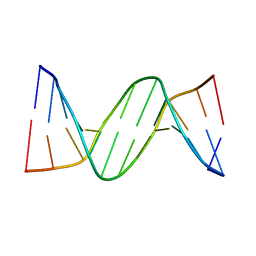 | | B-DNA with 7-deaza-dG modification | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*DTP*DCP*(7GU)P*DCP*DG)-3') | | Authors: | Wang, F, Li, F, Ganguly, M, Marky, L.A, Gold, B, Egli, M, Stone, M.P. | | Deposit date: | 2007-06-25 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A bridging water anchors the tethered 5-(3-aminopropyl)-2'-deoxyuridine amine in the DNA major groove proximate to the N+2 C.G base pair: implications for formation of interstrand 5'-GNC-3' cross-links by nitrogen mustards.
Biochemistry, 47, 2008
|
|
2QEF
 
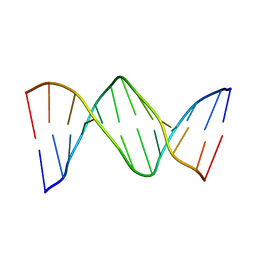 | | X-ray structure of 7-deaza-dG and Z3dU modified duplex CGCGAATXCZCG | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*(ZDU)P*DCP*(7GU)P*DCP*DG)-3') | | Authors: | Wang, F, Li, F, Ganguly, M, Marky, L.A, Gold, B, Egli, M, Stone, M.P. | | Deposit date: | 2007-06-25 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A bridging water anchors the tethered 5-(3-aminopropyl)-2'-deoxyuridine amine in the DNA major groove proximate to the N+2 C.G base pair: implications for formation of interstrand 5'-GNC-3' cross-links by nitrogen mustards.
Biochemistry, 47, 2008
|
|
2PFC
 
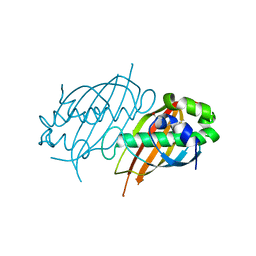 | | Structure of Mycobacterium tuberculosis Rv0098 | | Descriptor: | Hypothetical protein Rv0098/MT0107, PALMITIC ACID | | Authors: | Wang, F, Sacchettini, J.C. | | Deposit date: | 2007-04-04 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a type III thioesterase reveals the function of an operon crucial for Mtb virulence.
Chem.Biol., 14, 2007
|
|
6EF8
 
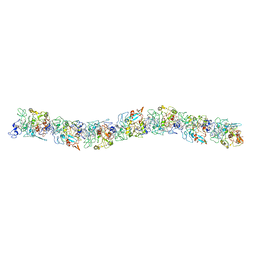 | | Cryo-EM of the OmcS nanowires from Geobacter sulfurreducens | | Descriptor: | C-type cytochrome OmcS, HEME C | | Authors: | Wang, F, Gu, Y, Egelman, E.H, Malvankar, N.S. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-10 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers.
Cell, 177, 2019
|
|
6JWD
 
 | | structure of RET G-quadruplex in complex with berberine | | Descriptor: | BERBERINE, DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6JWE
 
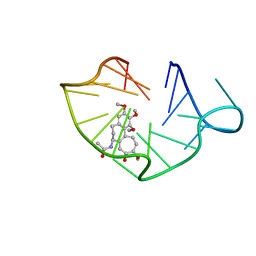 | | structure of RET G-quadruplex in complex with colchicine | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-20 | | Release date: | 2020-02-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
