3SRK
 
 | | A new class of suicide inhibitor blocks nucleotide binding to pyruvate kinase | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, POTASSIUM ION, Pyruvate kinase, ... | | Authors: | Morgan, H.P, Walsh, M, Blackburn, E.A, Wear, M.A, Boxer, M, Shen, M, McNae, I.W, Michels, P.A.M, Auld, D.S, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A new class of suicide inhibitor blocks nucleotide binding to pyruvate kinase
To be Published
|
|
4YW4
 
 | | Streptococcus pneumoniae sialidase NanC | | Descriptor: | GLYCEROL, Neuraminidase C, PHOSPHATE ION | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a novel sialidase
To be published
|
|
3BJH
 
 | | Soft-SAD crystal structure of a pheromone binding protein from the honeybee Apis mellifera L. | | Descriptor: | GLYCEROL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Lartigue, A, Gruez, A, Briand, L, Blon, F, Bezirard, V, Walsh, M, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sulfur single-wavelength anomalous diffraction crystal structure of a pheromone-binding protein from the honeybee Apis mellifera L.
J.Biol.Chem., 279, 2004
|
|
1DY9
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor I) | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1DY8
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor II) | | Descriptor: | N-[(benzyloxy)carbonyl]-L-isoleucyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70) | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1DXP
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (apo structure) | | Descriptor: | GLYCEROL, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-13 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1SFK
 
 | | Core (C) protein from West Nile Virus, subtype Kunjin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Core protein, ... | | Authors: | Dokland, T, Walsh, M, Mackenzie, J.M, Khromykh, A.A, Ee, K.-H, Wang, S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | West nile virus core protein; tetramer structure and ribbon formation
Structure, 12, 2004
|
|
1UW7
 
 | | Nsp9 protein from SARS-coronavirus. | | Descriptor: | NSP9 | | Authors: | Sutton, G, Fry, E, Carter, L, Sainsbury, S, Walter, T, Nettleship, J, Berrow, N, Owens, R, Gilbert, R, Davidson, A, Siddell, S, Poon, L.L.M, Diprose, J, Alderton, D, Walsh, M, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Nsp9 Replicase Protein of Sars-Coronavirus, Structure and Functional Insights
Structure, 12, 2004
|
|
8POA
 
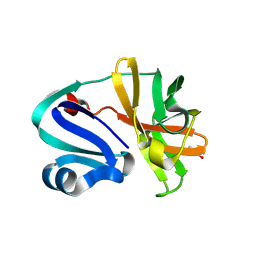 | | Structure of Coxsackievirus A16 (G-10) 2A protease | | Descriptor: | GLYCEROL, Protease 2A, ZINC ION | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-04 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7PMO
 
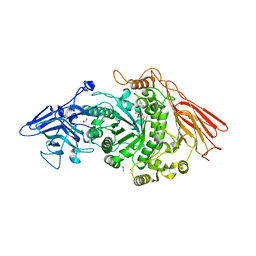 | | Ruminococcus gnavus ATC29149 endo-beta-1,4-galactosidase (RgGH98) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Owen, C.D, Wu, H, Crost, E.H, van Bakel, W, Gascuena, A.M, Latousakis, D, Hicks, T, Walpole, S, Urbanowicz, P.A, Ndeh, D, Monaco, S, Salom, L.S, Griffiths, R, Colvile, A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2021-09-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.
Plos Biol., 19, 2021
|
|
7Q20
 
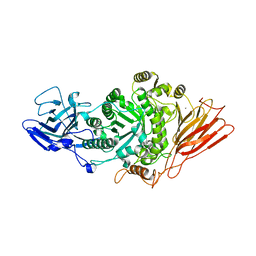 | | Ruminococcus gnavus ATC29149 endo-beta-1,4-galactosidase (RgGH98) in complex with blood group A trisaccharide | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Ruminococcus gnavus endogalactosidase GH98, ... | | Authors: | Owen, C.D, Wu, H, Crost, E.H, van Bakel, W, Gascuena, A.M, Latousakis, D, Hicks, T, Walpole, S, Urbanowicz, P.A, Ndeh, D, Monaco, S, Salom, L.S, Griffiths, R, Colvile, A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2021-10-22 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.
Plos Biol., 19, 2021
|
|
7Q1W
 
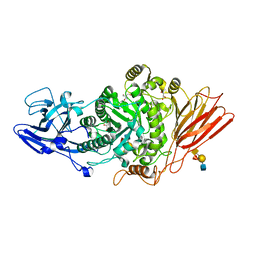 | | Ruminococcus gnavus ATC29149 endo-beta-1,4-galactosidase (RgGH98) E411A in complex with blood group A (BgA II) tetrasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Owen, C.D, Wu, H, Crost, E.H, van Bakel, W, Gascuena, A.M, Latousakis, D, Hicks, T, Walpole, S, Urbanowicz, P.A, Ndeh, D, Monaco, S, Salom, L.S, Griffiths, R, Colvile, A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.
Plos Biol., 19, 2021
|
|
5FRK
 
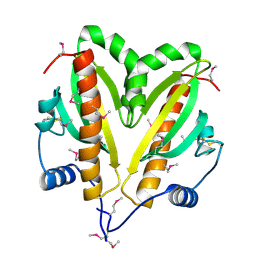 | | SeMet crystal structure of Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | Descriptor: | AMYR | | Authors: | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | Deposit date: | 2015-12-18 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
5FR7
 
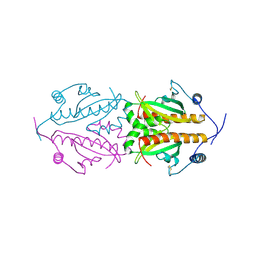 | | Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | Descriptor: | AMYR | | Authors: | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | Deposit date: | 2015-12-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
4B7F
 
 | | Structure of a liganded bacterial catalase | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Gumiero, A, Walsh, M. | | Deposit date: | 2012-08-20 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Structural and Dynamic Investigation of the Inhibition of Catalase by Nitric Oxide.
Org.Biomol.Chem., 11, 2013
|
|
4YW5
 
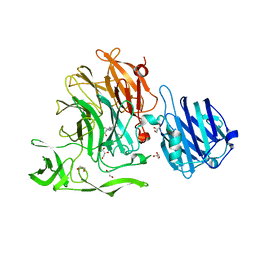 | | Crystal Structure of Streptococcus pneumoniae NanC, complex with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4YW1
 
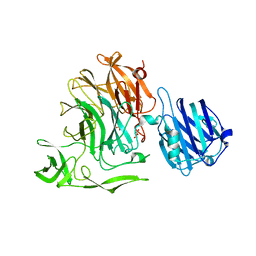 | | Crystal Structure of Streptococcus pneumoniae NanC, complex with Neu5Ac and Neu5Ac2en following soaking with 3'SL | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4YW3
 
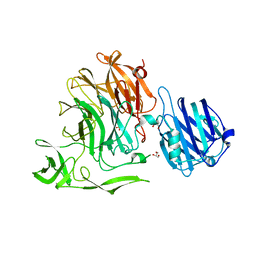 | | Crystal Structure of Streptococcus pneumoniae NanC, complex with Neu5Ac and Neu5Ac2en following soaking with Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
5MF5
 
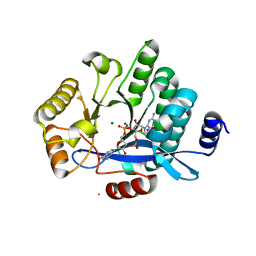 | | PA3825-EAL Mg-CdG Structure | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Diguanylate phosphodiesterase, MAGNESIUM ION | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2016-11-17 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
1W3C
 
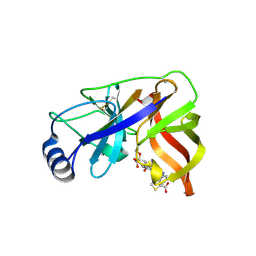 | | Crystal structure of the Hepatitis C Virus NS3 Protease in complex with a peptidomimetic inhibitor | | Descriptor: | 3-({(2S)-2-[({(1R)-1-[({(1R)-1-[(R)-CARBOXY(HYDROXY)METHYL]-3,3-DIFLUOROPROPYL}AMINO)CARBONYL]-3-METHYLBUTYL}AMINO)CARB ONYL]-2,3-DIHYDRO-1H-INDOL-2-YL}METHYL)THIOPHENE-2-CARBOXYLIC ACID, 3-({(2S)-2-[({(1S)-1-[({(1S)-1-[(R)-CARBOXY(HYDROXY)METHYL]-3,3-DIFLUOROPROPYL}AMINO)CARBONYL]-3-METHYLBUTYL}AMINO)CARBONYL]-2,3-DIHYDRO-1H-INDOL-2-YL}METHYL)THIOPHENE-2-CARBOXYLIC ACID, NONSTRUCTURAL PROTEIN NS4A (P4), ... | | Authors: | Di Marco, S, Volpari, C. | | Deposit date: | 2004-07-14 | | Release date: | 2004-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Design and Enzyme-Bound Crystal Structure of Indoline Based Peptidomimetic Inhibitors of Hepatitis C Virus Ns3 Protease
J.Med.Chem., 47, 2004
|
|
5F9T
 
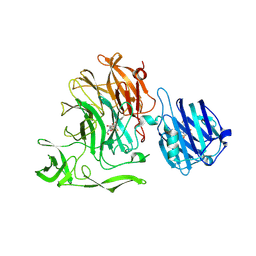 | | Crystal Structure of Streptococcus pneumoniae NanC, covalent complex with a fluorinated Neu5Ac derivative | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, GLYCEROL, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-12-10 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4YW2
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, complex 6'SL | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4B7G
 
 | | Structure of a bacterial catalase | | Descriptor: | CATALASE, CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gumiero, A, Walsh, M. | | Deposit date: | 2012-08-20 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Structural and Dynamic Investigation of the Inhibition of Catalase by Nitric Oxide.
Org.Biomol.Chem., 11, 2013
|
|
4B7H
 
 | | Structure of a highdose liganded bacterial catalase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CATALASE, CHLORIDE ION, ... | | Authors: | Gumiero, A, Walsh, M. | | Deposit date: | 2012-08-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Structural and Dynamic Investigation of the Inhibition of Catalase by Nitric Oxide.
Org.Biomol.Chem., 11, 2013
|
|
1C7F
 
 | | D95E OXIDIZED FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | McCarthy, A, Walsh, M, Higgins, T, D'Arcy, D. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Investigation of the Role of Aspartate 95 in the Modulation of the Redox Potentials Of Desulfovibrio Vulgaris Flavodoxin
Biochemistry, 41, 2002
|
|
