4E2I
 
 | | The Complex Structure of the SV40 Helicase Large T Antigen and p68 Subunit of DNA Polymerase Alpha-Primase | | Descriptor: | DNA polymerase alpha subunit B, Large T antigen, ZINC ION | | Authors: | Zhou, B, Arnett, D.R, Yu, X, Brewster, A, Sowd, G.A, Xie, C.L, Vila, S, Gai, D, Fanning, E, Chen, X.S. | | Deposit date: | 2012-03-08 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Structural basis for the interaction of a hexameric replicative helicase with the regulatory subunit of human DNA polymerase alpha-primase.
J.Biol.Chem., 287, 2012
|
|
6EHT
 
 | | Modulation of PCNA sliding surface by p15PAF suggests a suppressive mechanism for cisplatin-induced DNA lesion bypass by pol eta holoenzyme | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), PCNA-associated factor, ... | | Authors: | De March, M, Barrera-Vilarmau, S, Mentegari, E, Merino, N, Bressan, E, Maga, G, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
6GIS
 
 | | Structural basis of human clamp sliding on DNA | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Barrera-Vilarmau, S, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis of human PCNA sliding on DNA.
Nat Commun, 8, 2017
|
|
6FJY
 
 | | Crystal structure of CsuC-CsuE chaperone-tip adhesion subunit pre-assembly complex from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuC, Protein CsuE | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-16 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis forAcinetobacter baumanniibiofilm formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5D6H
 
 | | Crystal structure of CsuC-CsuA/B chaperone-major subunit pre-assembly complex from Csu biofilm-mediating pili of Acinetobacter baumannii | | Descriptor: | CsuA/B, CsuC | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight into Archaic and Alternative Chaperone-Usher Pathways Reveals a Novel Mechanism of Pilus Biogenesis.
Plos Pathog., 11, 2015
|
|
6FM5
 
 | | Crystal structure of self-complemented CsuA/B major subunit from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B,CsuA/B,CsuA/B | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
6FQ0
 
 | | Crystal structure of the CsuC-CsuA/B chaperone-subunit preassembly complex of the archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B, CsuC | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-02-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
6FQA
 
 | | Crystal structure of the CsuC-CsuA/B chaperone-subunit preassembly complex of the archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B, CsuC | | Authors: | Parilova, O, Pakharukova, N.A, Malmi, H, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-02-13 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
2L5B
 
 | |
2L58
 
 | |
3ETP
 
 | | The crystal structure of the ligand-binding domain of the EphB2 receptor at 2.0 A resolution | | Descriptor: | Ephrin type-B receptor 2 | | Authors: | Goldgur, Y, Paavilainen, S, Nikolov, D.B, Himanen, J.P. | | Deposit date: | 2008-10-08 | | Release date: | 2008-10-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the ligand-binding domain of the EphB2 receptor at 2 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7ZL4
 
 | | Cryo-EM structure of archaic chaperone-usher Csu pilus of Acinetobacter baumannii | | Descriptor: | CsuA/B | | Authors: | Pakharukova, N, Malmi, H, Tuittila, M, Paavilainen, S, Ghosal, D, Chang, Y.W, Jensen, G.J, Zavialov, A.V. | | Deposit date: | 2022-04-13 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Archaic chaperone-usher pili self-secrete into superelastic zigzag springs.
Nature, 609, 2022
|
|
9C50
 
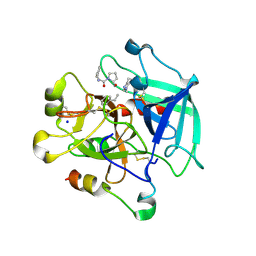 | | Replacement of a single residue changes the primary specificity of thrombin | | Descriptor: | FPF, SODIUM ION, Thrombin A-chain, ... | | Authors: | Dei Rossi, A, Deavila, S, Mohammed, B.M, Korolev, S, Di Cera, E. | | Deposit date: | 2024-06-05 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Replacement of a single residue changes the primary specificity of thrombin.
J.Thromb.Haemost., 2025
|
|
6GWS
 
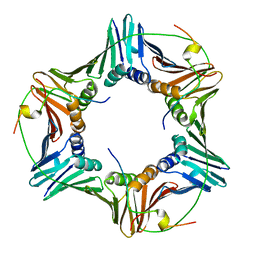 | | Crystal structure of human PCNA in complex with three p15 peptides | | Descriptor: | PCNA-associated factor, Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Gonzalez-Magana, A, Romano-Moreno, M, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
5LN8
 
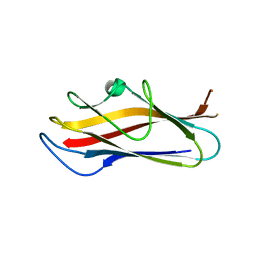 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica, in complex with galactose | | Descriptor: | Fimbrial protein MyfA,Fimbrial protein MyfA, beta-D-galactopyranose | | Authors: | Pakharukova, N.A, Roy, S, Rahman, M.M, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LND
 
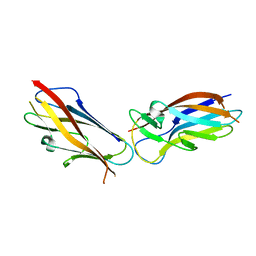 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica | | Descriptor: | Fimbrial protein MyfA,Fimbrial protein MyfA,Fimbrial protein MyfA | | Authors: | Pakharukova, N.A, Roy, S, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-04 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LN4
 
 | | Crystal structure of self-complemented PsaA, the major subunit of pH 6 antigen from Yersinia pests, in complex with choline | | Descriptor: | CHOLINE ION, pH 6 antigen,pH 6 antigen | | Authors: | Pakharukova, N.A, Roy, S, Rahman, M.M, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LO7
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Fimbrial protein MyfA,Fimbrial protein MyfA | | Authors: | Pakharukova, N.A, Roy, S, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5KTG
 
 | |
2NAQ
 
 | | 3D NMR solution structure of NLRP3 PYD | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | de Alba, E, Oroz, J. | | Deposit date: | 2016-01-07 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ASC Pyrin Domain Self-associates and Binds NLRP3 Protein Using Equivalent Binding Interfaces.
J.Biol.Chem., 291, 2016
|
|
7M69
 
 | |
7M68
 
 | |
1QOX
 
 | |
5DFK
 
 | |
3TTY
 
 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus in complex with galactose | | Descriptor: | Beta-galactosidase, ZINC ION, alpha-D-galactopyranose | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
