8U0Q
 
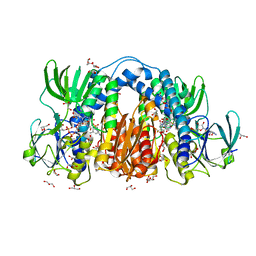 | | Co-crystal structure of optimized analog TDI-13537 provided new insights into the potency determinants of the sulfonamide inhibitor series | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dementiev, A.A, Michino, M, Vendome, J, Ginn, J, Bryk, R, Olland, A. | | Deposit date: | 2023-08-29 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Shape-Based Virtual Screening of a Billion-Compound Library Identifies Mycobacterial Lipoamide Dehydrogenase Inhibitors.
Acs Bio Med Chem Au, 3, 2023
|
|
6UKJ
 
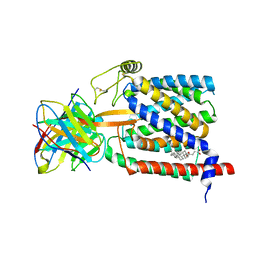 | | Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Chloroquine resistance transporter, Fab Heavy Chain, ... | | Authors: | Kim, J, Tan, Y.Z, Wicht, K.J, Erramilli, S.K, Dhingra, S.K, Okombo, J, Vendome, J, Hagenah, L.M, Giacometti, S.I, Warren, A.L, Nosol, K, Roepe, P.D, Potter, C.S, Carragher, B, Kossiakoff, A.A, Quick, M, Fidock, D.A, Mancia, F. | | Deposit date: | 2019-10-05 | | Release date: | 2019-12-04 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and drug resistance of the Plasmodium falciparum transporter PfCRT.
Nature, 576, 2019
|
|
4AKT
 
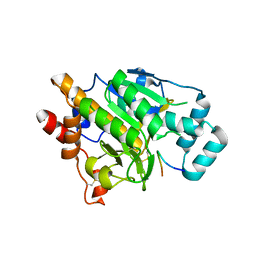 | | PatG macrocyclase in complex with peptide | | Descriptor: | SUBSTRATE ANALOGUE, THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | Authors: | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-18 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AKS
 
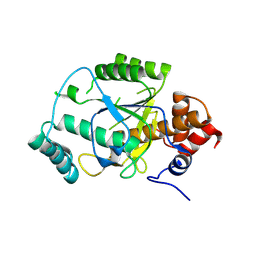 | | PatG macrocyclase domain | | Descriptor: | THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | Authors: | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-18 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3ZXX
 
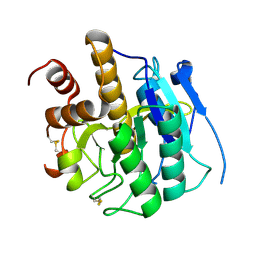 | | Structure of self-cleaved protease domain of PatA | | Descriptor: | SUBTILISIN-LIKE PROTEIN | | Authors: | Koehnke, J, Zollman, D, Vendome, J, Raab, A, Houssen, W.E, Smith, M.C, Jaspars, M, Naismith, J.H. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Discovery of New Cyanobactins from Cyanothece Pcc 7425 Defines a New Signature for Processing of Patellamides.
Chembiochem, 13, 2012
|
|
3ZXY
 
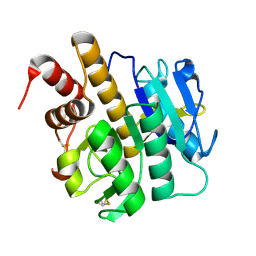 | | Structure of S218A mutant of the protease domain of PatA | | Descriptor: | SUBTILISIN-LIKE PROTEIN | | Authors: | Koehnke, J, Zollman, D, Vendome, J, Raab, A, Houssen, W.E, Smith, M.C, Jaspars, M, Naismith, J.H. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Discovery of New Cyanobactins from Cyanothece Pcc 7425 Defines a New Signature for Processing of Patellamides.
Chembiochem, 13, 2012
|
|
4NQQ
 
 | |
4NUP
 
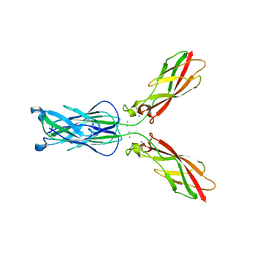 | |
4NUM
 
 | | Crystal structure of mouse N-cadherin EC1-2 A78SI92M | | Descriptor: | CALCIUM ION, Cadherin-2 | | Authors: | Jin, X. | | Deposit date: | 2013-12-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and energetic determinants of adhesive binding specificity in type I cadherins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NUQ
 
 | | Crystal structure of mouse N-cadherin EC1-2 W2F | | Descriptor: | CALCIUM ION, Cadherin-2 | | Authors: | Jin, X. | | Deposit date: | 2013-12-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Structural and energetic determinants of adhesive binding specificity in type I cadherins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3QRB
 
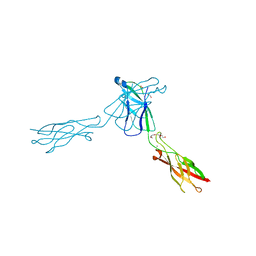 | | crystal structure of E-cadherin EC1-2 P5A P6A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cadherin-1, ... | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2011-02-17 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular design principles underlying beta-strand swapping in the adhesive dimerization of cadherins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6WNK
 
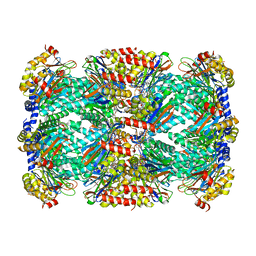 | | Macrocyclic peptides TDI5575 that selectively inhibit the Mycobacterium tuberculosis proteasome | | Descriptor: | (12S,15S)-N-[(2-fluorophenyl)methyl]-10,13-dioxo-12-{2-oxo-2-[(2R)-2-phenylpyrrolidin-1-yl]ethyl}-2-oxa-11,14-diazatricyclo[15.2.2.1~3,7~]docosa-1(19),3(22),4,6,17,20-hexaene-15-carboxamide, CITRIC ACID, DIMETHYLFORMAMIDE, ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Macrocyclic Peptides that Selectively Inhibit the Mycobacterium tuberculosis Proteasome.
J.Med.Chem., 64, 2021
|
|
5XUR
 
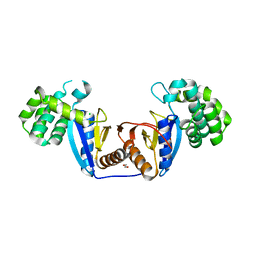 | | Crystal Structure of Rv2466c C22S Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Thioredoxin-like reductase Rv2466c | | Authors: | Zhang, X, Li, H. | | Deposit date: | 2017-06-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Identification of a Mycothiol-Dependent Nitroreductase from Mycobacterium tuberculosis.
ACS Infect Dis, 4, 2018
|
|
7N8L
 
 | |
7N8E
 
 | |
7N8M
 
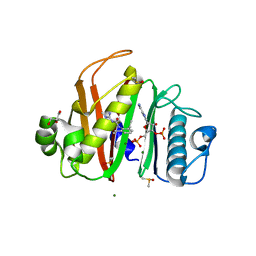 | |
5LHK
 
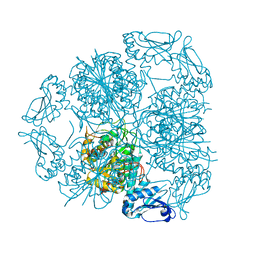 | | Bottromycin maturation enzyme BotP in complex with Mn | | Descriptor: | BICARBONATE ION, Leucine aminopeptidase 2, chloroplastic, ... | | Authors: | Koehnke, J, Adam, S. | | Deposit date: | 2016-07-12 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Substrate Recognition of the Bottromycin Maturation Enzyme BotP.
Chembiochem, 17, 2016
|
|
5LHJ
 
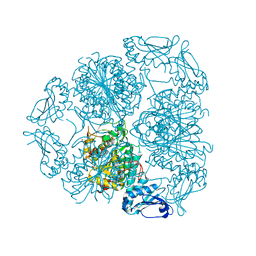 | | Bottromycin maturation enzyme BotP | | Descriptor: | CHLORIDE ION, GLYCEROL, Leucine aminopeptidase 2, ... | | Authors: | Koehnke, J, Mann, G. | | Deposit date: | 2016-07-12 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and Substrate Recognition of the Bottromycin Maturation Enzyme BotP.
Chembiochem, 17, 2016
|
|
5F15
 
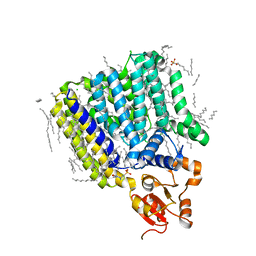 | | Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose (L-Ara4N) transferase, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
5EZM
 
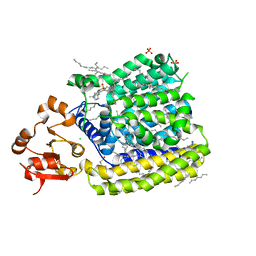 | | Crystal Structure of ArnT from Cupriavidus metallidurans in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferases of PMT family, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
3Q2L
 
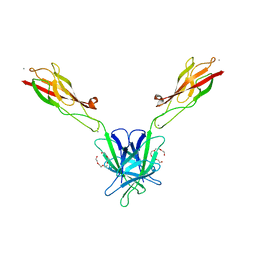 | | Mouse E-cadherin EC1-2 V81D mutant | | Descriptor: | CALCIUM ION, Cadherin-1, PENTAETHYLENE GLYCOL | | Authors: | Harrison, O.J, Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
3UBG
 
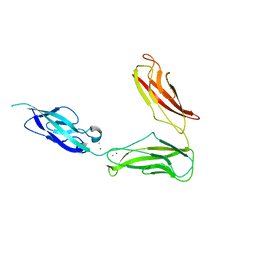 | | Crystal structure of Drosophila N-cadherin EC1-3, II | | Descriptor: | CALCIUM ION, Neural-cadherin, ZINC ION | | Authors: | Jin, X, Walker, M.A, Shapiro, L. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structures of Drosophila N-cadherin ectodomain regions reveal a widely used class of Ca2+-free interdomain linkers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3Q2N
 
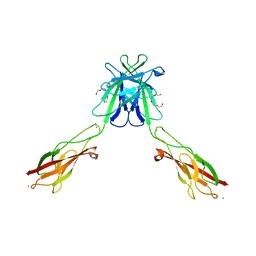 | | Mouse E-cadherin EC1-2 L175D mutant | | Descriptor: | CALCIUM ION, Cadherin-1, TETRAETHYLENE GLYCOL | | Authors: | Harrison, O.J, Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
3Q2V
 
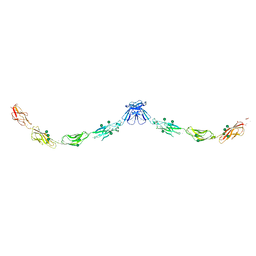 | | Crystal structure of mouse E-cadherin ectodomain | | Descriptor: | CALCIUM ION, Cadherin-1, MANGANESE (II) ION, ... | | Authors: | Jin, X, Harrison, O.J, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
3UBF
 
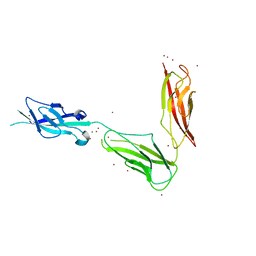 | | Crystal structure of Drosophila N-cadherin EC1-3, I | | Descriptor: | CALCIUM ION, Neural-cadherin, ZINC ION | | Authors: | Jin, X, Walker, M.A, Shapiro, L. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Drosophila N-cadherin ectodomain regions reveal a widely used class of Ca2+-free interdomain linkers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
