6AU2
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4H,6H-[1,2,4]triazolo[4,3-a][4,1]benzoxazepine, BETA-MERCAPTOETHANOL, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-10-11 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification and characterization of the first fragment hits for SETDB1 Tudor domain.
Bioorg.Med.Chem., 27, 2019
|
|
6MF9
 
 | | Crystal structure of CGD4-650 with compound BI2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, ZnKn (C2HC)+Athook+bromo domain protein, Taf250, ... | | Authors: | Dong, A, Lin, Y.L, Hou, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-10 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | Crystal structure of CGD4-650 with compound BI2536
to be published
|
|
1U1Z
 
 | | The Structure of (3R)-hydroxyacyl-ACP dehydratase (FabZ) | | Descriptor: | (3R)-hydroxymyristoyl-[acyl carrier protein] dehydratase, SULFATE ION | | Authors: | Kimber, M.S, Martin, F, Lu, Y, Houston, S, Vedadi, M, Dharamsi, A, Fiebig, K.M, Schmid, M, Rock, C.O. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa
J.Biol.Chem., 279, 2004
|
|
1XG5
 
 | | Structure of human putative dehydrogenase MGC4172 in complex with NADP | | Descriptor: | ACETIC ACID, ARPG836, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kavanagh, K, Ng, S, Sharma, S, Vedadi, M, von Delft, F, Walker, J.R, dhe Paganon, S, Bray, J, Oppermann, U, Edwards, A, Arrowsmith, C, Sundstrom, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-09-16 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Genomics Consortium: Structure of the putative human dehydrogenase MGC4172
To be Published
|
|
6NEY
 
 | | Crystal structure of TcBDF5, a bromodomain containing protein from Trypanosoma cruzi | | Descriptor: | Uncharacterized protein | | Authors: | Loppnau, P, Dong, A, Tempel, W, Lin, Y.H, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Vedadi, M, Harding, R.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-18 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of TcBDF5, a bromodomain containing protein from Trypanosoma cruzi
To Be Published
|
|
6NEZ
 
 | | Trypanosoma brucei - BDF5, Tb427tmp.01.5000 A, solved with PF-CBP1 | | Descriptor: | 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-[2-(4-propoxyphenyl)ethyl]-1H-benzimidazole, UNKNOWN ATOM OR ION, Uncharacterized protein | | Authors: | Lin, Y.H, Dong, A, Tempel, W, McAuley, J, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Vedadi, M, Harding, R.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-18 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trypanosoma brucei - BDF5, Tb427tmp.01.5000 A, solved with PF-CBP1
to be published
|
|
6NIM
 
 | | Trypanosoma cruzi - BDF2, TcCLB.506553.20, solved with bromosporine | | Descriptor: | Bromodomain factor 2 protein, Bromosporine, SULFATE ION, ... | | Authors: | Lin, Y.H, Dong, A, Tempel, W, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Vedadi, M, Harding, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Trypanosoma cruzi - BDF2, TcCLB.506553.20, solved with bromosporine
to be published
|
|
6NP7
 
 | | Crystal structure of Trypanosoma cruzi bromodomain BDF2 (TcCLB.506553.20) | | Descriptor: | Bromodomain factor 2 protein | | Authors: | Lin, Y.H, Dong, A, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Hui, R, Harding, R.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-17 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Trypanosoma cruzi bromodomain BDF2 (TcCLB.506553.20)
to be published
|
|
8BWU
 
 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Transcription factor ETV6,Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Konkolova, E, Klima, M, Boura, E, Jin, J, Kaniskan, H.U, Han, Y, Vedadi, M. | | Deposit date: | 2022-12-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Drug Discovery in Low Data Regimes: Leveraging a Computational Pipeline for the Discovery of Novel SARS-CoV-2 Nsp14-MTase Inhibitors.
Biorxiv, 2024
|
|
4JLG
 
 | | SETD7 in complex with inhibitor (R)-PFI-2 and S-adenosyl-methionine | | Descriptor: | 8-fluoro-N-{(2R)-1-oxo-1-(pyrrolidin-1-yl)-3-[3-(trifluoromethyl)phenyl]propan-2-yl}-1,2,3,4-tetrahydroisoquinoline-6-sulfonamide, Histone-lysine N-methyltransferase SETD7, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Wu, H, Zeng, H, El Bakkouri, M, Barsyte, D, Vedadi, M, Tatlock, J, Owen, D, Bunnage, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4IA9
 
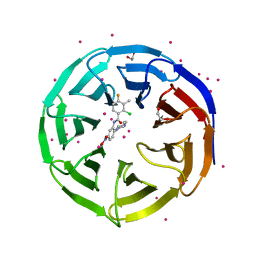 | | Crystal structure of human WD REPEAT DOMAIN 5 in complex with 2-chloro-4-fluoro-3-methyl-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-4-fluoro-3-methyl-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Bolshan, Y, Getlik, M, Tempel, W, Kuznetsova, E, Wasney, G.A, Hajian, T, Poda, G, Nguyen, K.T, Schapira, M, Brown, P.J, Al-awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Smil, D, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-06 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Synthesis, Optimization, and Evaluation of Novel Small Molecules as Antagonists of WDR5-MLL Interaction.
ACS Med Chem Lett, 4, 2013
|
|
4JDS
 
 | | SETD7 in complex with inhibitor PF-5426 and S-adenosyl-methionine | | Descriptor: | Histone-lysine N-methyltransferase SETD7, N-[(2R)-3-(3-cyanophenyl)-1-oxo-1-(pyrrolidin-1-yl)propan-2-yl]-8-fluoro-1,2,3,4-tetrahydroisoquinoline-6-sulfonamide, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Wu, H, Zeng, H, Park, H, El Bakkouri, M, Barsyte, D, Vedadi, M, Tatlock, J, Owen, D, Bunnage, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETD7 in complex with inhibitor PF-5426 and S-adenosyl-methionine
to be published
|
|
5EAP
 
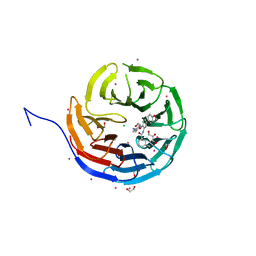 | | Crystal structure of human WDR5 in complex with compound 9e | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
5EAR
 
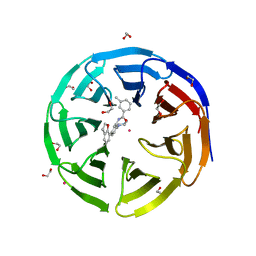 | | Crystal structure of human WDR5 in complex with compound 9d | | Descriptor: | 1,2-ETHANEDIOL, N-[5-(2,3-dihydro-1-benzofuran-7-yl)-2-(4-methylpiperazin-1-yl)phenyl]-3-methylbenzamide, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
5EAL
 
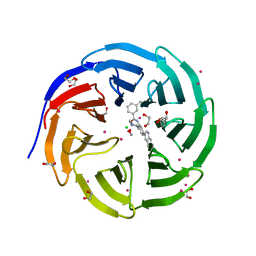 | | Crystal structure of human WDR5 in complex with compound 9h | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-quinolin-3-yl-phenyl]benzamide, CHLORIDE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human WDR5 in complex with compound 9h
to be published
|
|
5EAM
 
 | | Crystal structure of human WDR5 in complex with compound 9o | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
4NVQ
 
 | | Human G9a in Complex with Inhibitor A-366 | | Descriptor: | 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Sweis, R.F, Pliushchev, M, Brown, P.J, Guo, J, Li, F, Maag, D, Petros, A.M, Soni, N.B, Tse, C, Vedadi, M, Michaelides, M.R, Chiang, G.G, Pappano, W.N. | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and development of potent and selective inhibitors of histone methyltransferase g9a.
ACS Med Chem Lett, 5, 2014
|
|
6AU3
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, N-{[2-(3,5-dimethyl-4H-1,2,4-triazol-4-yl)phenyl]methyl}acetamide, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments
to be published
|
|
4EQZ
 
 | | Crystal structure of human DOT1L in complex with inhibitor FED2 | | Descriptor: | 5'-deoxy-5'-[(3-{[(4-methylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]adenosine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
4ER0
 
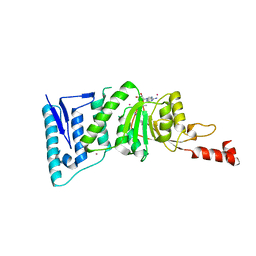 | | Crystal Structure of human DOT1L in complex with inhibitor FED1 | | Descriptor: | 5'-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5'-deoxyadenosine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
4ER7
 
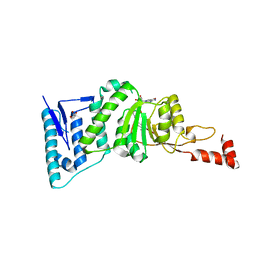 | | Crystal Structure of human DOT1L in complex with inhibitor SGC0947 | | Descriptor: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
3MA6
 
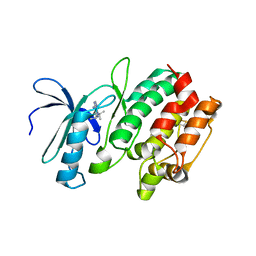 | | Crystal structure of kinase domain of TgCDPK1 in presence of 3BrB-PP1 | | Descriptor: | 3-(3-bromobenzyl)-1-tert-butyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1 | | Authors: | Wernimont, A.K, Qiu, W, Amani, M, Artz, J.D, Hassani, A.A, Senisterra, G, Vedadi, M, Sibley, L.D, Lourido, S, Shokat, K, Zhang, C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-07-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of kinase domain of TgCDPK1 in presence of 3BrB-PP1
To be Published
|
|
4HSG
 
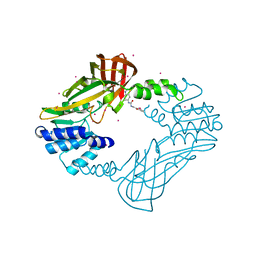 | | Crystal structure of human PRMT3 in complex with an allosteric inhibitor (PRMT3- KTD) | | Descriptor: | 1-(1,2,3-benzothiadiazol-6-yl)-3-(2-oxo-2-phenylethyl)urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dobrovetsky, E, Dong, A, Liu, F, Li, F, Tempel, W, Siarheyeva, A, Hajian, T, Smil, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting an allosteric binding site of PRMT3 yields potent and selective inhibitors.
J. Med. Chem., 56, 2013
|
|
3Q5I
 
 | | Crystal Structure of PBANKA_031420 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Panico, E, Crombet, L, Cossar, D, Hassani, A, Vedadi, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Neculai, A.M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of CDPK1 from Plasmodium Bergheii, PBANKA_031420
To be Published
|
|
5VAC
 
 | | Crystal Structure of ATXR5 SET domain in complex with K36me3 histone H3 peptide | | Descriptor: | DIMETHYL SULFOXIDE, Histone H3.2, Probable Histone-lysine N-methyltransferase ATXR5, ... | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
